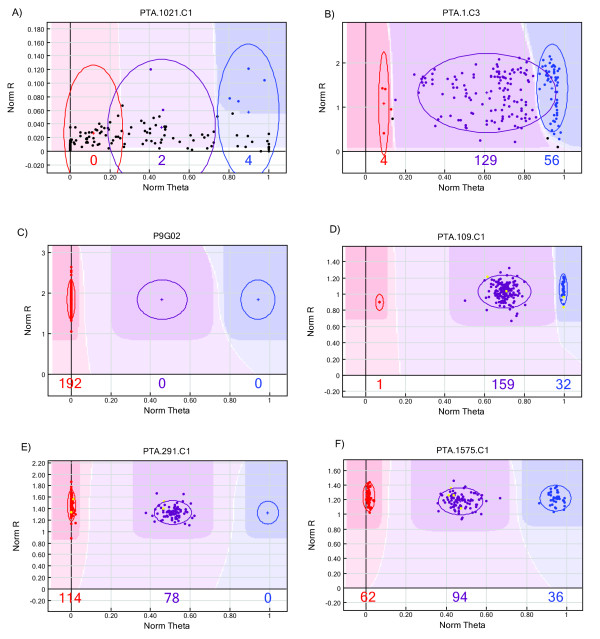
Figure 1.
Examples of SNP graphs observed in Lolium oligo pool assay (LOPA1) GoldenGate genotyping. SNP graphs are illustrated using the Software Illumina® GenomeStudio, version 2009.2. The normalized R (y-axis) is the normalized sum of intensities of the two dyes (Cy3 and Cy5), the normalized Theta (x-axis) is the deviation of Cy3 and Cy5 fluorescence from pure Cy3 and pure Cy5 signal (0 and 1). A normalized Theta value close to 0 and 1 is homozygous for SNP variant 1 and 2, respectively, a heterozygous sample is in between. The red, blue and purple ovals have the diameter of two standard deviations computed from the dispersal of the red, blue and purple dots, respectively. The numbers of plants in each cluster are indicated below the x-axis. (A) The 192 samples genotyped for SNP marker PTA.1021.C1 revealed fluorescence signal intensities close to 0, indicating assay failure. (B) Although the clustering algorithm at SNP PTA.1.C3 distinguished the three clusters at a GenTrain score of 0.40, such a genotyping pattern was considered inaccurate and this SNP was discarded from further analysis. (C) This illustration shows the SNP graph of monomorphic P9G02. (D) and (E) illustrate dominant SNPs being homozygous in one and heterozygous in the other mapping parent. For genetic linkage mapping, the markers PTA.109.C1 and PTA.291.C1 followed the segregation type nnxnp and lmxll, respectively [53]. Dots corresponding to the parents of the VrnA mapping population (which are represented in duplicates) are highlighted in yellow. Graph (F) shows a classical example of a SNP marker being heterozygous in both parents following the segregation pattern hkxhk.