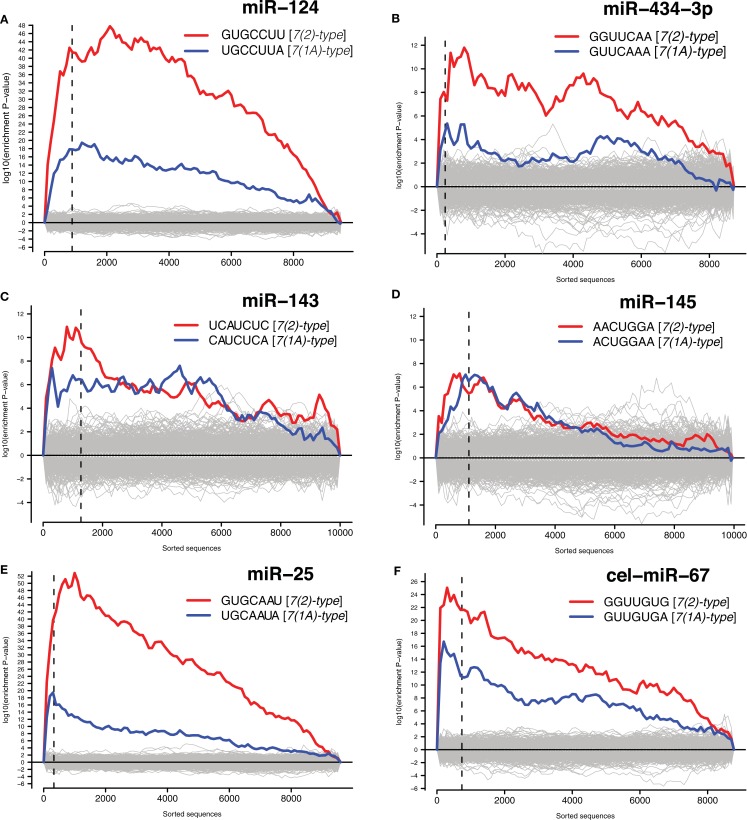
Figure 3.
Identified putative direct miRNA targets. The x-axes represent 3′UTRs corresponding to genes sorted from most downregulated to most upregulated in comparison to cultures transfected with (A) miR-124 mimic and inhibitor, (B) miR-434-3p mimic and inhibitor, (C) miR-143 mimic and inhibitor, (D) miR-145 mimic and inhibitor, (E) miR-25 mimic and inhibitor, (F) ce-miR-67 mimic and mock transfected cultures. Sequences to the left of the vertical dashed lines correspond to genes downregulated with differential expression adjusted P < 0.05. Putative targets of miRNAs are defined as downregulated genes (adjusted P < 0.05) encoding transcripts with one or 7(2) or 7(1A)-type seed matching sites (Bartel, 2009) in 3′UTRs. Section “Materials and Methods” for details.