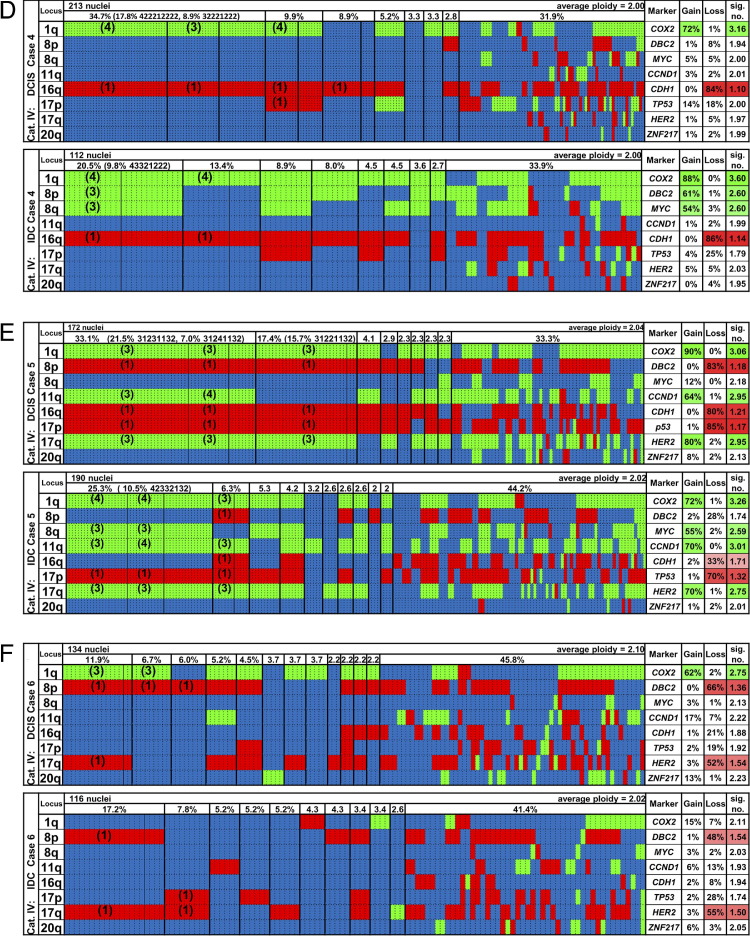
Figure 4.
Summary of imbalance clones in 6 of the 13 cases of DCIS and IDC (cases 1 to 6; A–F). Green, gains; red, losses; blue, unchanged. The organization of the graphs is the same for all cases and is explained for A (case 1) in detail from left to right. The “Locus” column shows the chromosome arm. Each vertical dotted line separates individual nuclei, and the vertical thin solid line discerns specific signal pattern clones among the most common imbalance clones, which are separated from each other by vertical thick solid lines. The row above the imbalance clones displays the percentages at which the clones were found (threshold ≥2%). For example, the DCIS of case 1 had 20% of cells with gains of COX2, DBC2, and MYC and a loss of CDH1, and the other markers were unchanged; among those cells, 15.8% had a signal pattern clone of 33421222 in the DCIS. In the IDC, 16.3% of cells had gains of COX2, DBC2, and MYC and a loss of CDH1, with no other changes observed; among those, 12.8% had a signal pattern clone of 33421222 (for the definitions of imbalance clone and signal pattern clone, see Materials and Methods). There were 8.4% of the cells with gains of COX2 and MYC and a loss of CDH1 as the only changes, and in this imbalance clone there were five cells with a signal pattern clone of 32421222. The “Marker” column shows the gene name. The “Gain” column shows that 51% of the cells had a gain of COX2, 36% of DBC2, and 60% of MYC in the DCIS. The “Loss” column shows that 55% of the cells had a loss of CDH1 in the DCIS. The “sig. no.” column shows that the average signal count for COX2 in the entire population was 2.53 (2.35 for DBC2, 3.06 for MYC, and 1.49 for CDH1). The percentage of gains or losses in >30% of the cells is also indicated by the color intensity. Ninety-five nuclei were counted for the DCIS of this case, and 86 for the IDC. Average ploidy values were calculated from the ploidy values assigned to each nucleus (see Materials and Methods). Columns that are presented in blue only had no copy number changes for any of the eight gene probes but signal numbers for centromere probes 4 and/or 10 that differed from the ploidy value. The remaining seven cases (cases 7 to 13) can be viewed in Supplemental Figure S1 (available at http://ajp.amjpathol.org).