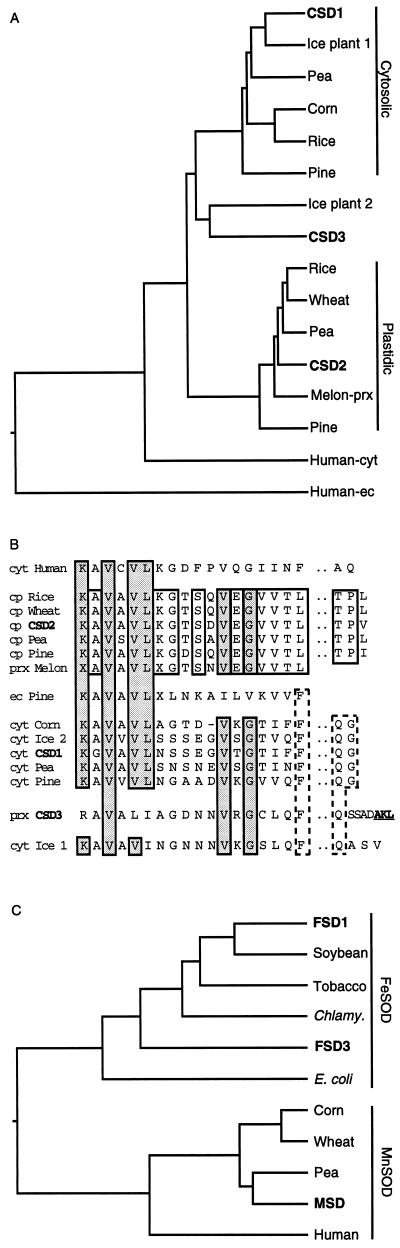
Figure 1.
Evolutionary relationships inferred from SOD sequences. Accession numbers for the sequences utilized are listed in Table II. A, Dendrogram of plant CuZnSOD amino acid sequences. The predicted amino acid sequences were analyzed utilizing a DNASTAR (Madison, WI) program, starting at Lys-Ala-Val-Ala-Val-Leu or a homologous sequence to eliminate any amino-terminal transit sequences. B, Alignment of the first 18 amino acids and conserved carboxy-terminal amino acids of the proteins used for the dendogram in A to illustrate cladistic relationships of the plant CuZnSODs. Underlined amino acids indicate a putative peroxisomal targeting sequence. prx, Peroxisome; cyt, cytosol; ec, extracellular. C, Dendogram of FeSOD and MnSOD amino acid sequences. The amino acid sequences were analyzed from the start of homology to the E. coli FeSOD sequence.