FIGURE 6:
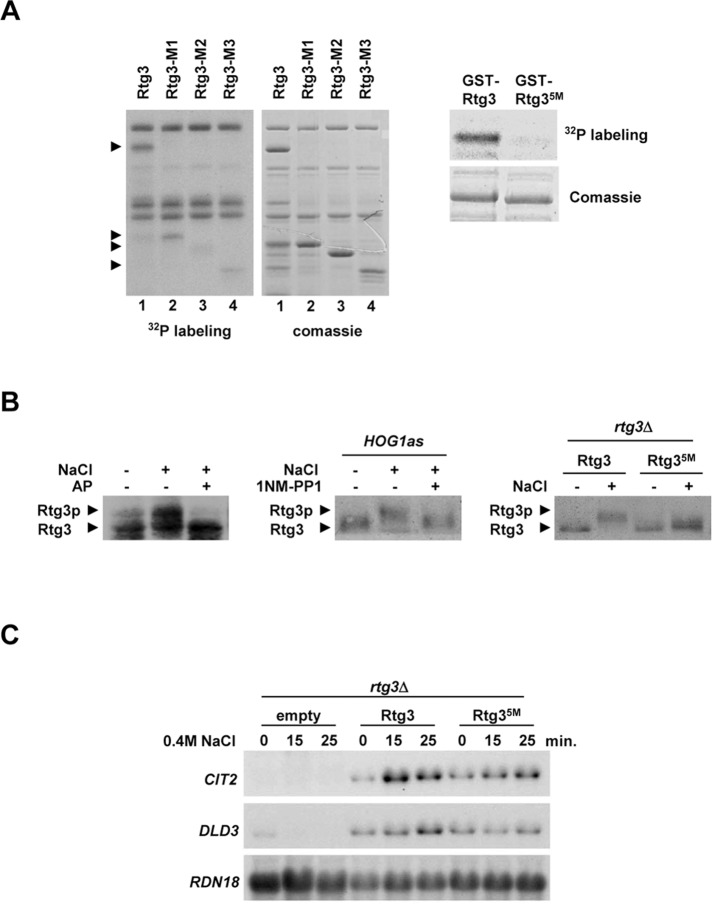
Rtg3 is phosphorylated by the Hog1 SAPK. (A) Hog1 phosphorylates Rtg3 in vitro. Left, GST-tagged full-length Rtg3 and fragments Rtg3-M1 (aa 1–210), Rtg3-M2 (aa 1–183), and Rtg3-M3 (aa 211–486) were purified from E. coli and assayed with in vitro activated Hog1. Phosphorylated proteins were detected by autoradiography, and proteins were detected by Coomassie Blue staining. Arrows indicate the positions of Rtg3 fragments. Right, mutations of Rtg3 Thr-197, Ser-222, Ser-227, Thr-249, and Ser-376 to Ala (Rtg35M) abolish in vitro phosphorylation by Hog1. (B) Rtg3 is phosphorylated upon osmostress. Left, rtg1Δ cells expressing Rtg3-HA were grown to 0.6–1 OD660, and samples were taken before (−) or 5 min after (+) addition of NaCl to a final concentration of 0.4 M. Protein extracts were treated (+) or not (−) with 10 U of AP. Rtg3-HA mobility was assessed by immunoblotting using anti-HA monoclonal antibody. Middle, rtg1Δhog1Δ cells expressing Rtg3-HA and carrying a mutant allele of HOG1 (HOG1as) were preincubated or not for 10 min with 5 μM of 1NM-PP1 prior to osmostress at the same conditions as in the left panel. Right, mutations of Rtg3 Thr-197, Ser-222, Ser-227, Thr-249, and Ser-376 to Ala (Rtg35M) abolish phosphorylation by osmostress. rtg1Δrtg3Δ strains were transformed with HA-tagged Rtg3 wild-type and the Rtg3 nonphosphorylatable mutant (Rtg35M) and processed as in the left panel. (C) Hog1 phosphorylation sites in Rtg3 are required for transcriptional activity. rtg3Δ strain was transformed with plasmids expressing wild-type Rtg3 or the Rtg3 nonphosphorylatable mutant (Rtg35M), grown in MD-Gln to 0.5–0.7 OD660, and subjected to osmotic stress (0.4 M NaCl) for the indicated length of time. Total RNA was assayed by Northern blot analysis for transcript levels of CIT2 and DLD3, with RDN18 as a loading control.