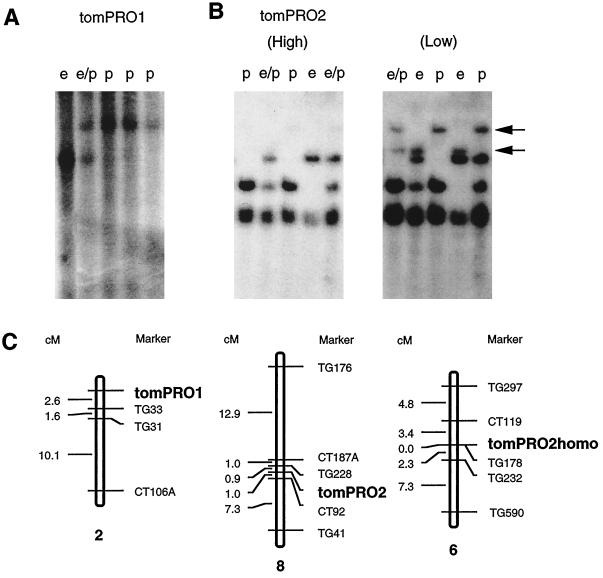
Figure 7.
Mapping of tomPRO1, tomPRO2, and tomPRO2 homolog in tomato nuclear genome. A and B, Southern-blot analyses of total DNA from the F2 population of crosses between L. esculentum and L. pennellii. Hybridizations were performed with the GK part of the tomPRO1 cDNA fragment (A) and the full length of tomPRO2 cDNA as the probes (B) at a high-stringency wash condition, 0.1× SSC, 0.1% SDS, at 42°C (High), and at a low-stringency wash condition, 0.2× SSC, 0.1% SDS, at 25°C (Low). Two bands that appeared at a low-stringency condition are depicted by arrows. This figure shows a representative portion of the blots, in which a total of 67 of the F2 populations were used for the RFLP mapping. Shown at the top of each lane is the RFLP pattern representative for the L. esculentum homozygote (e), the L. pennellii homozygote (p), or their heterozygote (e/p). C, Map position of tomPRO1, tomPRO2, and tomPRO2 homologs on the tomato chromosome. The maps were drawn by segregation analysis of RFLPs based on data by Tanksley et al. (1992). The map distances (in cM) are indicated on the left. Maps are not drawn to scale. tomPRO1, tomPRO2, and tomPRO2 homologs (tomPRO2homo) were located on chromosomes 2, 8, and 6, respectively.