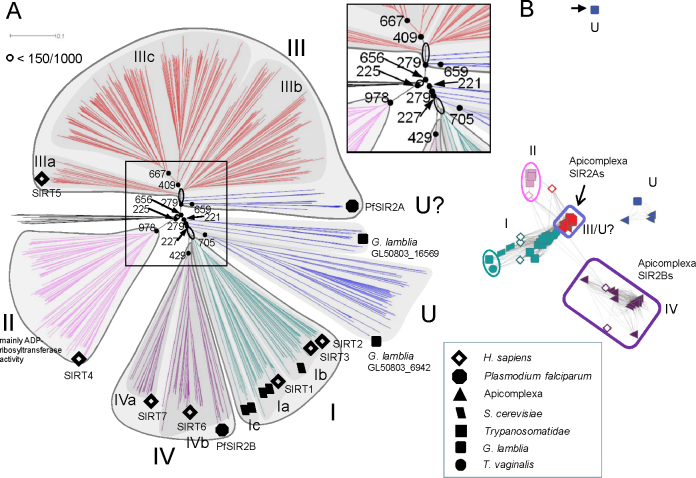
Fig. 1.
Sirtuin phylogeny with focus on parasitic protozoa. (A) A phylogenetic tree of 778 annotated sirtuins from different organisms (sources NCBI, EuPathDB, GeneDB) produced in ClustalX2 using Neighbour-Joining (NJ) method, with a bootstrap value of 1000. The phylogenetic tree branches were coloured according to the previous classification into 5 main clades: class I–IV (teal, pink, red and purple, respectively) and class U (blue) [10]. Note that several blue branches including Plasmodium SIR2As are assigned to group III (red) (as previously described in e.g.[10,12,13]) though with weak support. Therefore these sirtuins could also be grouped with class U sirtuins. Human sirtuin family members are marked with full diamonds. Incomplete protein sequences were removed from the analysis to facilitate the phylogenetic tree construction (for a list of sirtuins see Suppl. Table 1). The inset shows an enlargement clarification of the bootstrap values supporting particular nodes. (B) A Sequence Similarity Network (SSN) for the parasitic protozoa extracted from a global SSN built on BLAST pairwise alignments of sirtuin sequences from (A), with each node representing a single sirtuin and BLAST e-values as edges (cut-off threshold e15). The resulting SSN network intuitively represents the sirtuins class divisions (same colours as in B). Sirtuins of the parasitic protozoa contribute to every class of sirtuins. Apicomplexa sirtuins (triangles) belong mainly to class III/U and IV sirtuins (see text for details). H. sapiens (diamonds) and S. cerevisiae (parallelograms) SIRs are shown, and cluster accordingly to the previous classification [10]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)