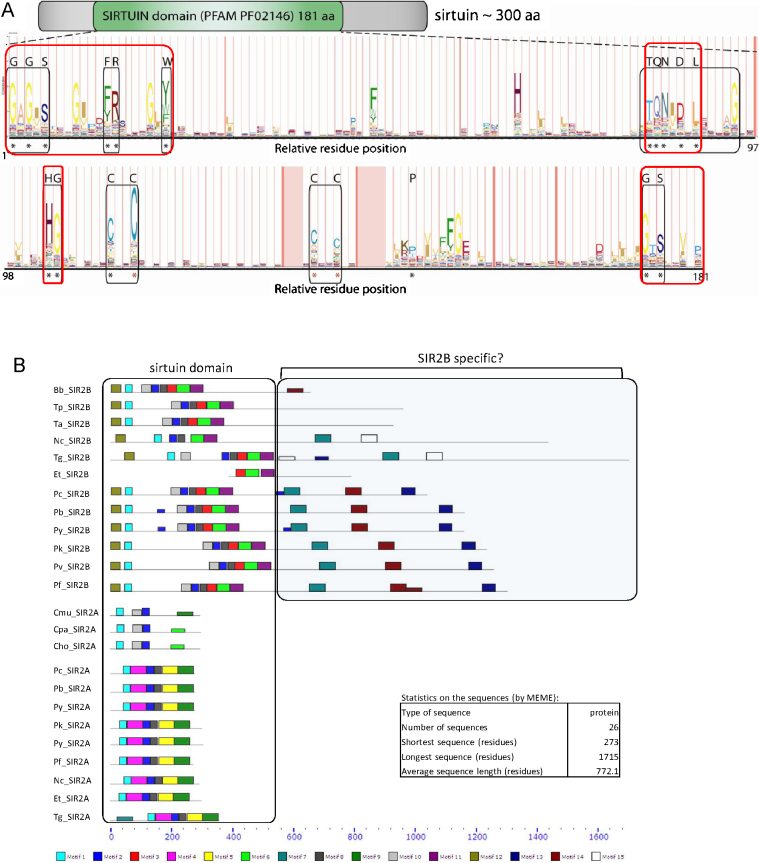
Fig. 2.
Structural organisation of the sirtuin domains of parasitic protozoa. (A) The average sirtuin of approximately 300 amino acids contains the canonical sirtuin domain (PFAM model PF02146) of 181 aa with several conserved regions (lower panel). Asterisks indicate residues conserved among parasitic protozoa according to ClustalX alignments (black = perfectly conserved, red = highly conserved). (B) Protein alignment of all Apicomplexa sirtuins with indicated subdomains characterised in silico by MEME sequence analysis tool (http://meme.sdsc.edu). The search was limited to 15 motifs/subdomains of between 6 and 50 aa. Sirtuin domain alignment (ClustalX2) with highlighted subdomains identified by MEME shows several highly conserved regions, with perfectly conserved residues within motif 1 (G[AS]GXS, FR), motif 2 (TQN[IV]D[SGN]L) and motif 8 (HG,CXXC). Motifs 7, 13 and 14 are present in most Apicomplexa SIR2B-like sirtuins, including all Plasmodium SIR2Bs, B. bovis SIR2B (Bb_SIR2B), N. caninum Nc_SIR2B, T. gondii Tg_SIR2B. Interestingly Cryptosporidium spp. sirtuins posesess some similarity in motif composition to Apicomplexa SIR2Bs. Note that Eimeria tenella Et_SIR2B sequence appears incomplete. Bb = Babesia bovis, Et = Eimeria tenella, Cryptosporidium: Cmu = C. muris, Cpa = C. parvum, Cho = C. hominis, Nc = Neospora caninum, Plasmodium: Pc = P. chabaudii, Pb = P. berghei, Py = P. yoelii, Pk = P. knowlesi, Pv = P. vivax, Pf = P. falciparum, Ta = Theileria annulata, Tp = Theileria parva, Tg = Toxoplasma gondii. (C) Top: 3D structure alignment of hSIRT5 and PfSIR2A (PDB IDs: 2B4Y gold and 3JWP blue respectively) using PyMOL (DeLano Scientific, www.pymol.org), structural alignment root mean squared (RMS) value = 2.0. PfSIR2A: The structure is shown with adenosine monophosphate (AMP) present in the NAD+-binding pocket in the Rossman fold (coloured red). hSIRT5: The structure is shown with adenosine-5-diphosphoribose (ADP) present in the NAD+-binding pocket in the Rossman fold (coloured green). The co-ordinated zinc ion is shown as grey (PfSIR2A) and black (hSIRT5) spheres. Bottom: 3D structure alignment of a class U sirtuin from Thermotoga maritima and PfSIR2A (PDB IDs: 3JR3 purple and 3JWP teal respectively). RMS value = 1.7. PfSIR2A: The structure is shown with adenosine monophosphate (AMP) present in the NAD+-binding pocket in the Rossman fold (coloured red). TmSIR2: The structure does not contain molecules in the NAD+-binding pocket. The co-ordinated zinc ion is shown as grey (PfSIR2A) and black (TmSIR2) spheres. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)