Figure 2.
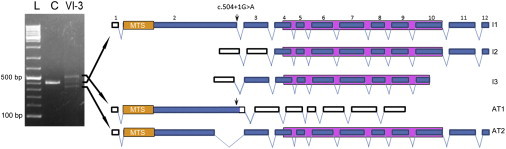
Schematic Representation of RMND1 Wild-Type and Aberrant Transcripts
RMND1 has four splicing variants, of which only three encode proteins: ENST00000367303 (12 exons, isoform 1 [I1]), ENST00000367303 (11 exons, isoform 2 [I2]), and ENST00000444024 (9 exons, isoform 3 [I3]). All three coding variants contain a DUF155 domain indicated by a purple box. Coding regions are represented by blue boxes, and noncoding regions are represented by white boxes. The longest transcript has a predicted N-terminal 33 amino acid mitochondrial targeting sequence (MTS). On the left is agarose gel showing the PCR product of RMND1 cDNA from control (C) and VI-3 fibroblasts. “L” indicates the 100 bp DNA ladder. Splice-site mutation c.504+1G>A (arrow) produces two aberrant transcripts (AT1 and AT2) and reduces the amount of wild-type transcript (middle band). The longer transcript, AT1, includes the insertion of 88 nucleotides after exon 2 and generates a premature stop at codon 171. In the shorter variant, AT2, a cryptic splice site in exon 2 is activated and results in an in-frame deletion of 75 nucleotides and a protein 25 amino acids shorter than the wild-type. RNA was extracted by the PureLink RNA Mini Kit (Ambion, Austin, TX) and reverse transcribed into cDNA with the VILO RT-PCR kit (Invitrogen, Grand Island, NY). RT-PCR of exons 2–7 was performed, and the RT-PCR products were electrophoresed in 2% agarose gels.
