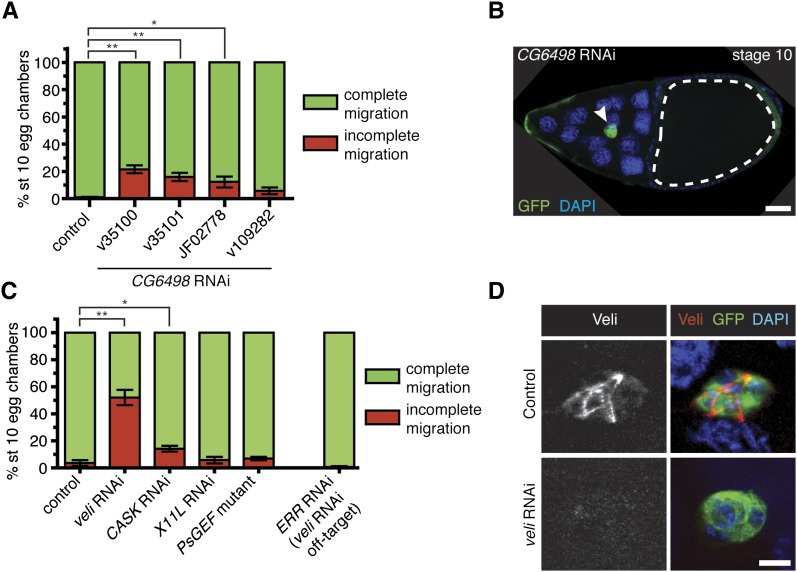
Figure 2 .
Confirmation of positive and negative hit genes. (A and C) Quantification of border cell migration at stage 10, shown as the percentage of border cells with complete (green) or incomplete (red) migration in egg chambers expressing RNAi to GFP (control) or the indicated RNAi transgenes driven by c306-GAL4. Error bars represent SEM; n ≥ 50 egg chambers in each of at least three trials (*P < 0.05; **P < 0.01; two-tailed unpaired t-test). (A) Knockdown of CG6498 using multiple transgenes disrupts border cell migration. (B) Representative example of an egg chamber with a border cell migration defect caused by CG6498 RNAi. Genotype is c306-GAL4/+; UAS-mCD8:GFP/UAS-CG6498 RNAi v35100. Border cells (green; arrowhead) stopped a little more than halfway to the oocyte (outlined). DAPI marks nuclei. Scale bar is 20 µm. (C) Border cell migration defects by RNAi knockdown of veli (v43094) and CASK (v34185). Normal border cell migration with RNAi knockdown of X11L (v28652) and in a PsGEF mutant (PsGEFΔ55/PsGEFΔ21). RNAi for ERR (line v108349), the predicted off-target gene for veli RNAi line v43094, did not disrupt border cell migration. (D) Border cells stained with an antibody to Veli. Control border cells (c306-GAL4/+; UAS-mCD8:GFP/+) had detectable Veli (red), which was strongly reduced in veli RNAi border cells (c306-GAL4/+; UAS-mCD8:GFP/UAS-veli RNAi v43094). GFP (green) shows GAL4 expression and DAPI (blue) labels nuclei. Scale bar is 10 µm.