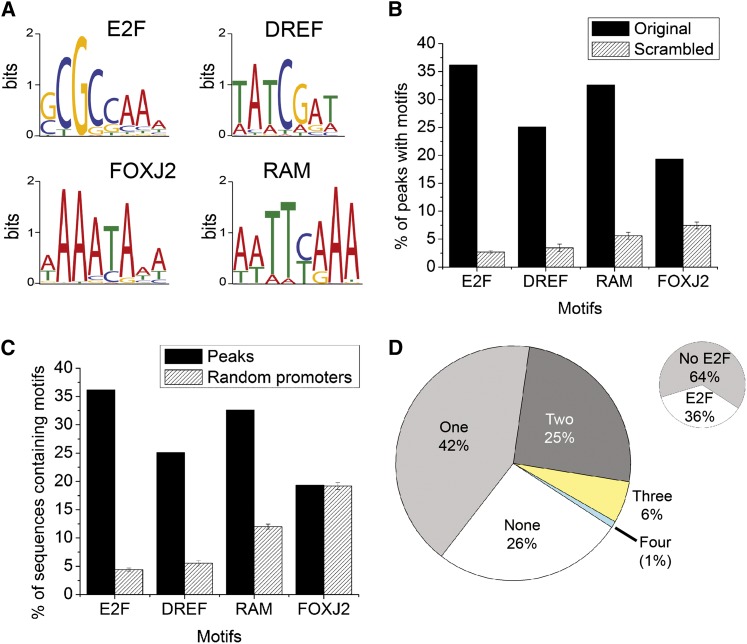
Figure 6 .
Transcription factor motifs enriched in Rbf1-bound peaks. (A) The four most overrepresented motifs identified by the MEME motif discovery tool, including one previously unknown motif (RAM). (B) Rbf1-associated motifs are highly enriched compared with average occurrence in DNA of the same A/T composition. The sequences under Rbf1 peaks were scrambled five times, and specific motifs with P < 0.0001 were identified. E2F sites showed the greatest level of enrichment in specifically bound regions compared with randomized DNA sequences. (C) E2F, DREF, and RAM motifs preferentially associate with Rbf1-bound promoters. The presence of motifs in Rbf1-bound sequences was compared with the presence in Rbf1-unbound promoters. FOXJ2 sites are not restricted to Rbf1-associated promoters and may represent a motif for a broadly acting factor. Note that the canonical DREF sites are 8-mers (Yamaguchi et al. 1995). In our data, the eighth nucleotide was not conserved. (D) Diversity of motif composition of peaks. A total of 42% of total peaks contained only one of the four different motifs (E2F, DREF, FOXJ2, or RAM). A quarter of the peaks had a combination of two different motifs; 6%, a combination of three, and 1% contained all four motifs. Only 36% or peaks had an identifiable E2F motif (small insert). Strikingly, a quarter of the peaks did not have any of the four motifs. The heterogeneity of sequences in Rbf1-bound peaks suggests that E2F may not be the only transcription factor that recruits Rbf1 to target gene promoters. Peaks used in this analysis were drawn from the 1236 bound regions found in both Rbf1 ChIP biological experiments. A peak with multiple E2F motifs, but no other motif types, was counted as one type of motif; a similar treatment applies for the other three motifs. “None” means the peaks did not contain any motifs for E2F, DREF, FOXJ2, or RAM.