Fig. 1.
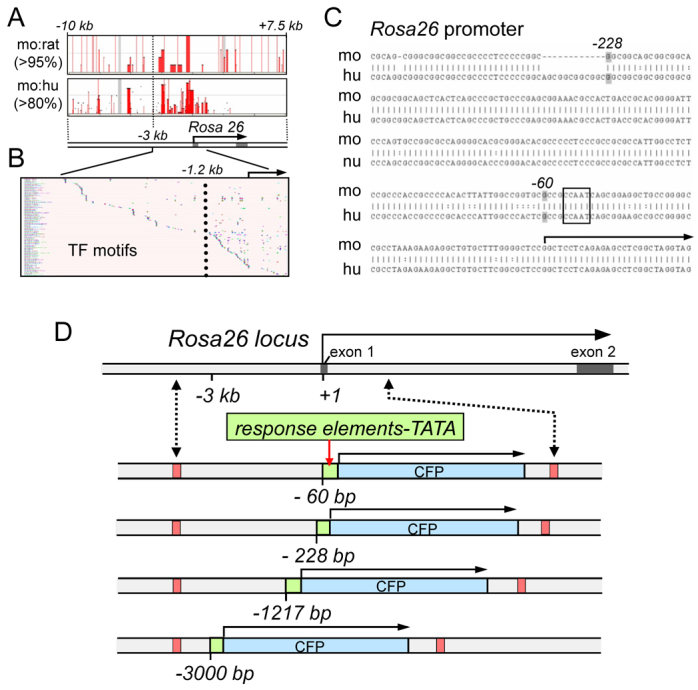
Rosa26 promoter analysis and design of deletions for signaling sentinel substitution. (A) Scheme depicts the Rosa26 locus; arrow, transcribed region. Boxes above depict sequence identities over 95% and 80% for mouse and rat (mo:ra) and mouse and human (mo:hu), respectively. Red bars within the boxes indicate conserved domains, largely extending to −3 kb of the start site of transcription. (B) VISTA plot of transcription factor motif distribution across the proximal 3 kb of the mouse Rosa26 promoter region. Vertical dotted line indicates a marked increase in motif density at −1.2 kb. (C) Comparison of the mouse and human Rosa26 promoter regions. A natural break occurs at −228 bp and a CCAAT box is evident downstream of −60 bp. (D) RMCE at the Rosa26 locus (upper scheme) results in substitutions that replace the regions from −4 kb to +1 kb with recombinant sequences where signaling response sentinels (green boxes) are inserted between deletion endpoints −60, −228, −1217 and −3000 and a TATA box fused to a coding sequence for CFP.
