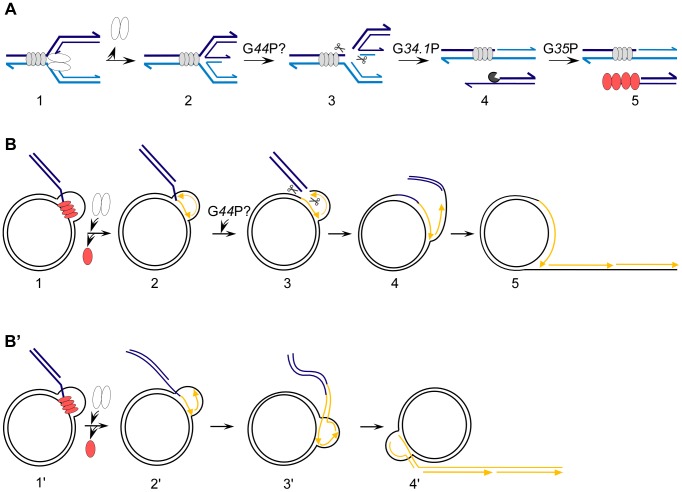
Figure 1. Putative roles of G44P in the generation of concatemeric linear SPP1 DNA after replication fork stalling.
(A) The generation of a 3′-tailed dsDNA molecule. 1, G38P (grey ovals) bound to oriR may hinder the progression of the replisome (white ovals), causing its subsequent disassembly. 2, The fork reverses and a Holliday junction (HJ) is formed. 3, The G44P HJ resolvase (scissors) may cleave this substrate and a one-ended double strand break is formed. 4, The G34.1P 5′-3′ exonuclease (pacman) processes the end. 5, The G35P recombinase (red ovals) forms filaments on the generated 3′-tailed duplex DNA. The 3′-ends of the strands are shown by arrows. From this substrate two alternatives were proposed to generate SPP1concatemeric DNA (B and B’). (B) Generation of concatemeric DNA by a sigma-like mechanism. 1, G35P promotes strand invasion on a supercoiled SPP1 molecule. 2, The replisome is recruited and the invaded strand primes DNA synthesis with subsequent dislodging of G35P. 3, G44P may cleave the strands of the replicated D-loop. 4 and 5, DNA synthesis followed by DNA ligation would generate the proper substrate for concatemeric DNA synthesis by a semiconservative mechanism. (B’) Bubble migration model for the generation of concatemeric DNA. 1′ and 2′, These steps are common in both avenues. 3′ and 4′, The replication bubble migrates and the newly synthesized strands are extruded, so that by this mechanism a concatemer is formed by conservative DNA synthesis, without an obvious need for a D-loop resolvase. In B and B’ the template DNA is drawn in black and newly synthesized DNA in yellow. The steps where G44P could participate are indicated by the scissors.