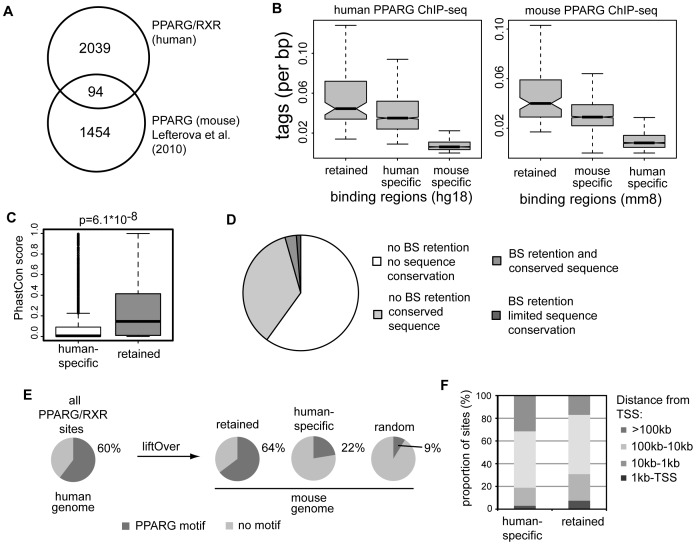
Figure 2. PPARG binding is poorly conserved between human and mouse macrophages.
A) Overlap of PPARG bindings sites between human and mouse macrophages. Comparison is based on murine binding sites lifted over to the human genome. 1548 out of 1961 PPARG binding sites in mouse aligned to the human genome. B) Tag counts from human PPARG library at different genomic loci in the human genome and mouse genome. Retained binding sites, human-specific sites and mouse-specific binding sites. Mouse and Human-specific sites in the human and mouse genome refer to the orthologous loci of mouse-specific or human-specific sites in the original genomes. For better visualization outliers were omitted from plot. C) Sequence conservation at human-specific and retained PPARG/RXR sites. Shown is the distribution of PhastCons scores for both categories. Significance was calculated using two-tailed t-test D) Pie chart summarizing the proportion of PPARG/RXR site that are retained and/or show sequence conservation (i.e. overlap with PhastCons element). E) Proportion of PPARG/RXR sites in human macrophages containing a PPARG motif compared to the proportion of sites with motif after liftOver to the mouse genome. The orthologous regions in the mouse genome are separated into PPARG bound and not bound. ‘Random’ shows the expected motif frequency for randomly distributed intervals with a matched size distribution. F) Distribution of PPARG/RXR binding sites in regard to TSS of RefGenes. Displayed are the distributions of human-specific and conserved PPARG/RXR sites.