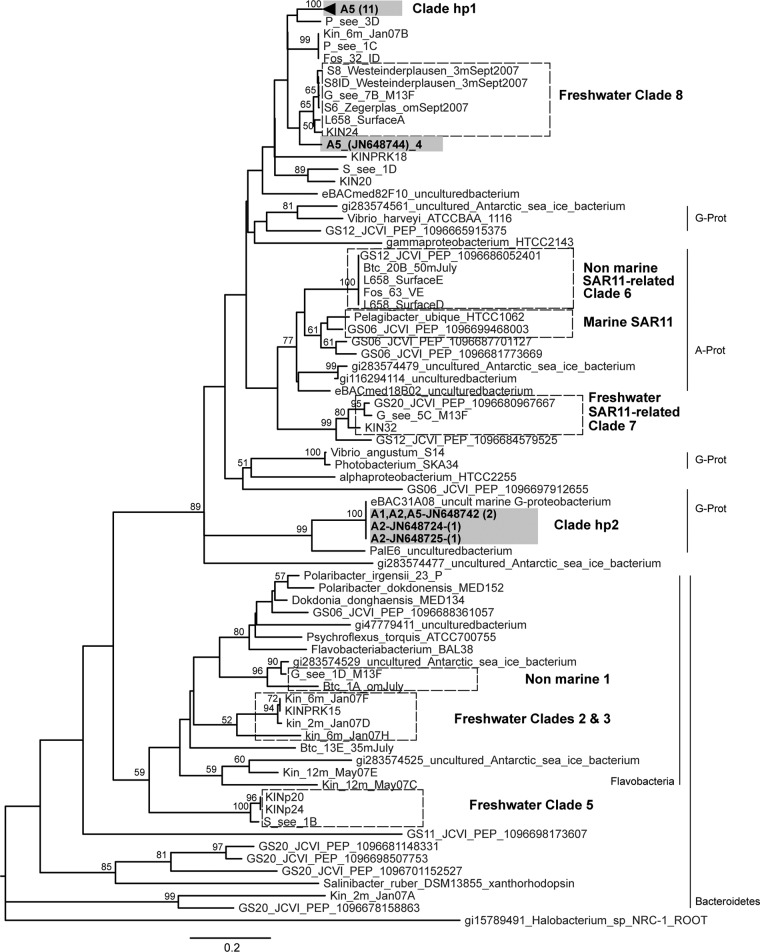
Fig 1.
Neighbor-joining phylogenetic tree constructed using 15 inferred PR protein hot spring sequences and 70 reported sequences. Sequences were aligned using ClustalW, and the phylogeny was constructed using the neighbor-joining algorithm with 1,000 bootstrap replicates and the program MEGA (24). Sequences from hot spring samples are shown in gray boxes, indicating the site (A1, A2, or A5) followed by the accession number and number of represented different nucleotide sequences (in parentheses). The number in parentheses for clade hp1 indicates the unique PR amino acid sequences. Hot spring clades (hp) and previously reported clades (hatched boxes) (2) are indicated. Bootstrap values greater than 50% are shown. Known phylogenetic affiliations are indicated. G-Prot, Gammaproteobacteria; A-Prot, Alphaproteobacteria.