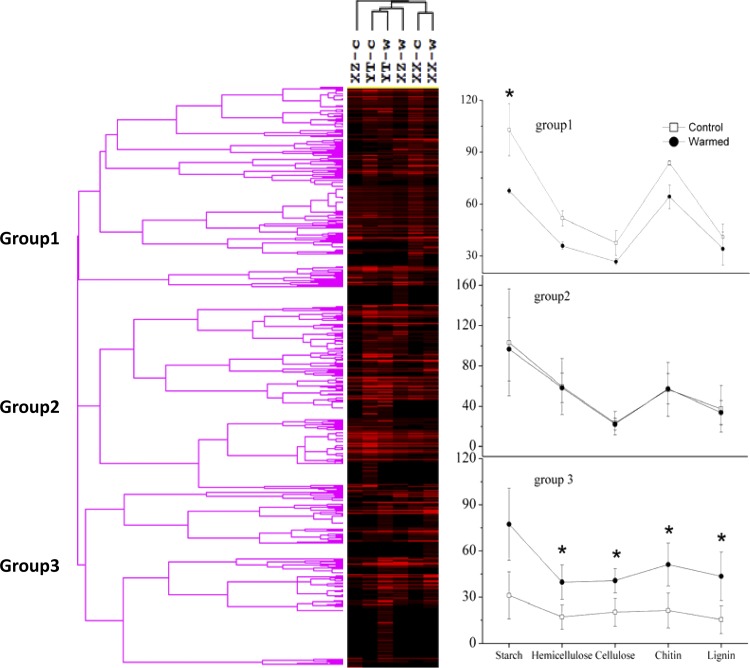
Fig 5.
Hierarchical cluster analysis of all C-degrading genes involved in carbon cycling by GeoChip 4.0. Results were generated in CLUSTER, version 3.0, using the Spearman rank correlation and the complete linkage method and visualized using TREEVIEW. Gray indicates signal intensities above background while black indicates signal intensities below background. Brighter red indicates higher signal intensities. YT-c, XZ-c, and XX-c represent control samples, and YT-w, XZ-w, and XX-w represent warmed samples. The abundance of detected genes, indicated by normalized signal intensities, involved in the degradation of different organic carbon pools (i.e., starch, hemicellulose, cellulose, chitin, and lignin) was plotted in three separate groups (group 1, group 2, and group 3). Genes clustered in group 3 showed a significant (P < 0.05) difference in abundance of detected genes between warmed and control samples. Error bars are ±1 standard deviation. Asterisks represent significant Student's t test differences between warmed and control samples (*, P < 0.05; **, P < 0.01).