Fig 1.
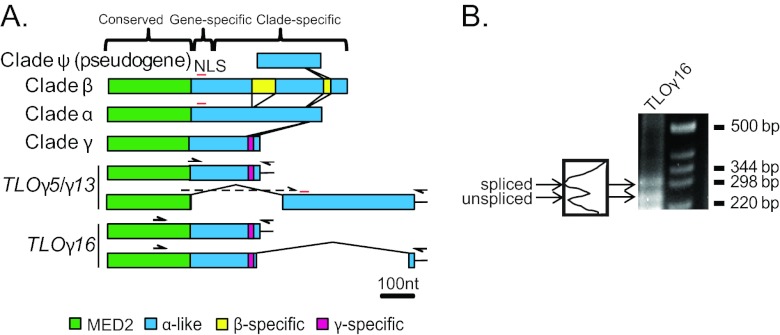
The C. albicans TLO genes form three separate clades. (A) Alignment of the TLO gene family members in the SC5314 genome and distinctions between the α, β, and γ clades. The TLO DNA sequence is roughly divided into three regions: a conserved region with the Med2 domain, highlighted in green; a gene-specific region consisting of highly variable adenosine-rich sequences; and a clade-specific region, which in the single-exon form is dissimilar between clades and highly conserved among genes within clades. A predicted nuclear localization signal (NLS) is indicated by a red line. A series of indels differentiates the TLOα/β and single-exon TLOγ sequences. Insertions of clade-specific sequences are connected by lines to indicate their positions relative to one another and color coded as indicated. Two differentially spliced transcripts were detected for TLOγ5 and TLOγ13 using a primer spanning the splicing event and against the poly(A) tail, indicated as half arrows. Sequencing confirmed splicing at nt 310 in TLOγ8 and at nt 333 in TLOγ13, in both cases joined to a second exon of ∼372 bp starting at nt 647 and 639, respectively. TLOγ16 splicing occurred 30 bp upstream of the stop codon in the unspliced transcript and spliced into a second, 45-bp exon from nt 356 to 401 downstream of the unspliced stop codon. (B) 3′ RACE of SC5314 cDNA amplified both spliced and unspliced transcripts of TLOγ16 of approximately 300 bp and 250 bp, respectively. Each transcript is expressed at roughly equivalent amounts as determined by densitometry.