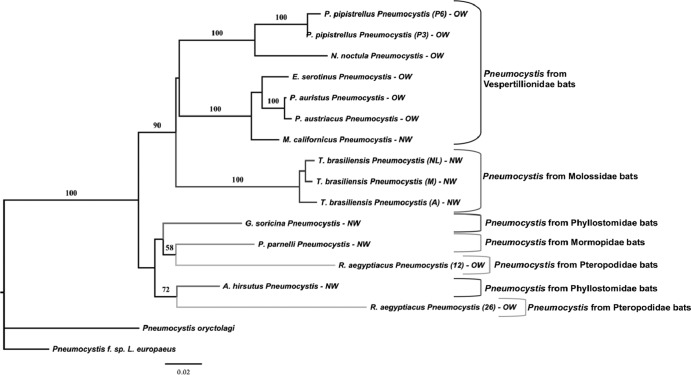
Fig 3.
Phenetic relationships of Pneumocystis organisms from bat species inferred from mtSSU rDNA sequences. The phylogram presented resulted from bootstrapped data sets obtained by using BIONJ analysis (heuristic search option in PAUP 4.0). The percentages above the branches are the frequencies with which a given branch appeared in 1,000 bootstrap replications. Bootstrap values below 50% are not displayed. Branch lengths correspond to the total nucleotide changes assigned to each branch by PAUP 4.0. Pneumocystis from rabbit (P. oryctolagi [47]) and from hare (P. f. sp. Lepus europaeus, GenBank accession JF431106) were chosen as outgroups. Letter and number codes indicate geographic origin and reference number of samples, respectively. NL, Nuevo León, Mexico; M, Michoacán, Mexico; A, Argentina; OW, Old World; NW, New World. Letters and numbers in parentheses indicate a particular sample (P3, P6, 12, 26).