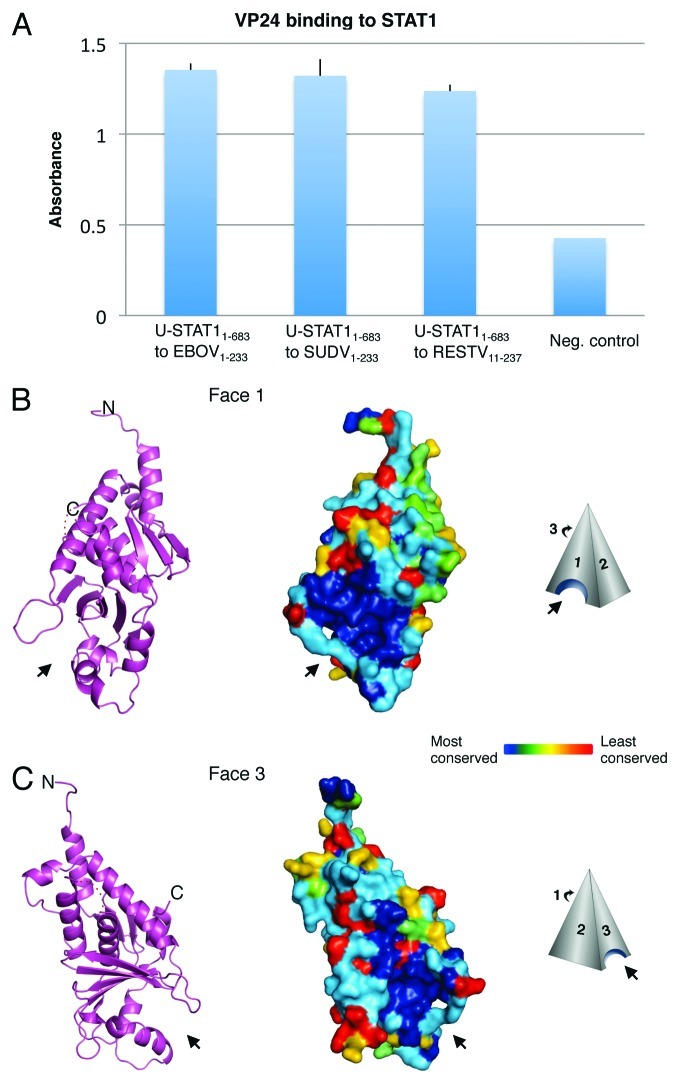
Figure 1. VP24 key features. (A) Purified EBOV1–233 VP24, SUDV1–233 VP24 and RESTV11–237 VP24 bind to purified STAT11–683 by ELISA. VP24s were coated onto the ELISA plate at 0.02 mg/ml for overnight incubation at room temperature. STAT11–683 was incubated on the plates, binding was detected with HRP-conjugated secondary antibody and the resulting O.D. was read at 450 nm. BSA was used as a negative control. (B and C) The overall shape of VP24 resembles a three-sided pyramid with Faces 1 and 3 as illustrated. A hydrophobic cavity in Face 1 and a polar cavity in Face 3 are indicated by arrows. A ribbon model of SUDV1–233 VP24 illustrates the underlying secondary structural elements, while a surface representation of SUDV1–233 VP24 indicates the sequence conservation. Navy indicates residues that are absolutely conserved among the ebola- and marburgviruses, while red indicates residues that are least conserved among these viruses. Face 2 is the least conserved of the three faces and is not illustrated. Sequence identity is calculated using Homolmapper.37 The accession codes for SUDV1–233, SUDV11–233 and RESTV11–237 are 3VNE, 3VNF and 4D9O, respectively.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.