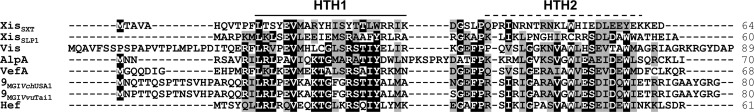
Fig 3.
Sequence alignment of the translation products of cds9MGIVvuTai1 and cds9MGIVchUSA1 with related RDFs. The primary sequences of RDFs encoded by two sequenced MGIs were aligned using MUSCLE with the transcriptional regulator AlpA from CP4-57 prophage (NP_417113) and the RDFs Xis encoded by ICEs of the SXT/R391 family (ACV96240), Hef of HPI from Y. pseudotuberculosis (CAB46594), VefA of VPI-2 from V. cholerae N16961 (NP_231420), SLP1 of plasmid SLP1 from Streptomyces coelicolor, and Vis of the satellite bacteriophage P4 (NP_042041). Amino acid residues that are identical or similar (BLOSUM62 substitution matrix) in at least 60% of the sequences are indicated by a black or gray background, respectively. The solid bar indicates a helix-turn-helix (HTH) DNA-binding motif predicted in all proteins based on the Dodd-Egan method (17) (Dodd-Egan scores of 2.36 or higher), whereas the dashed bar highlights a secondary HTH motif exclusively predicted in RDFs encoded by MGIs (Dodd-Egan score of 3.07). The length of each protein is indicated in the right column.