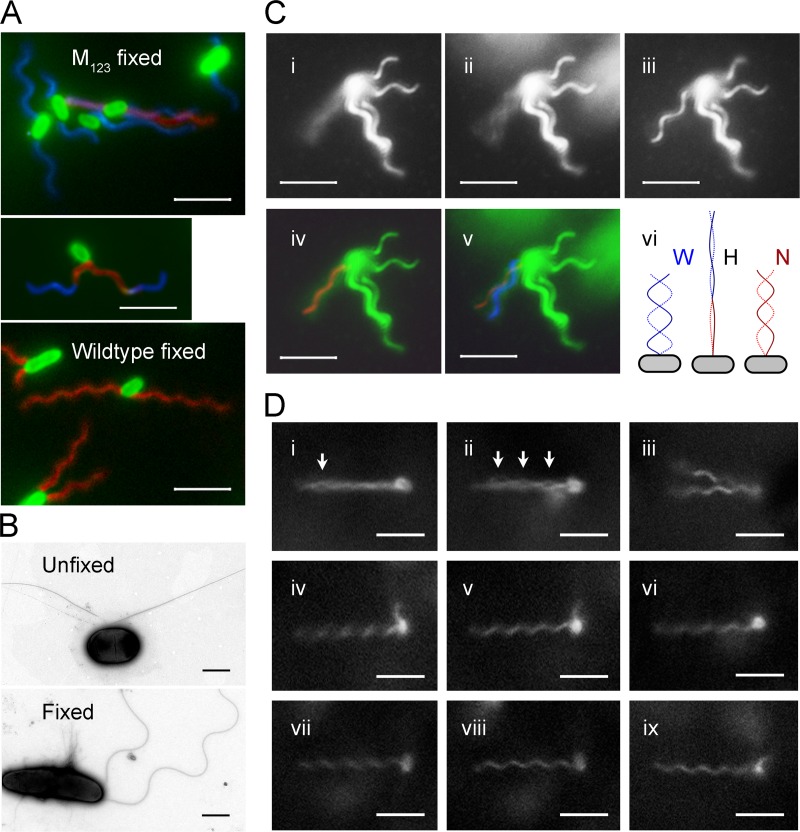
Fig 2.
Polymorphic transition of mutated flagella. The scale bar indicates 5 μm for fluorescence images and 1 μm for TEM images. (A) Fluorescence-labeled M123 cells (top) and wild-type cells (bottom) after fixation. Flagella are false-colored: red for normal form, blue for right-handed forms, and magenta for near-straight filament and transition segments. The middle panel shows a M123 cell with flagella undergoing polymorphic transition. (B) TEM images of negative-stained unfixed (top) and fixed (bottom) M123 cells. (C) Polymorphic transition of a flagellum on a tethered M123 cell at pH 8.0. Snapshots of a flagellum that was rotating fast (i), slowing down (ii), and stopped (iii) are on the top. The contrast was enhanced to show the flagellar image. (iv) Fitting a normal helix into the image cylinder of the rotating flagellum. (v) The w-coil helix formed as the rotation slowed down. (vi) Diagram showing the transformation of a w-coil helix during counterclockwise rotation. At the beginning (left), the flagellum is w-coil (W); when the threshold speed reached (middle), the flagella became hyperextended (H). Finally, the normal helix (N) is formed (right). (D) Snapshots of a M23 cell. (i) The reversion of a filament produced an eye in the bundle (arrow) when the swimming cell slowed down. (ii) More eyes formed before stop. (iii) Both flagella reverted to R-normal when stopped. (iv) One flagellum began to rotate. (v) Normal flagellum. (vi) Rotation reversed. (vii) Rotating clockwise. (viii) R-normal flagellum. (ix) Flagellum undergoing a polymorphic transition.