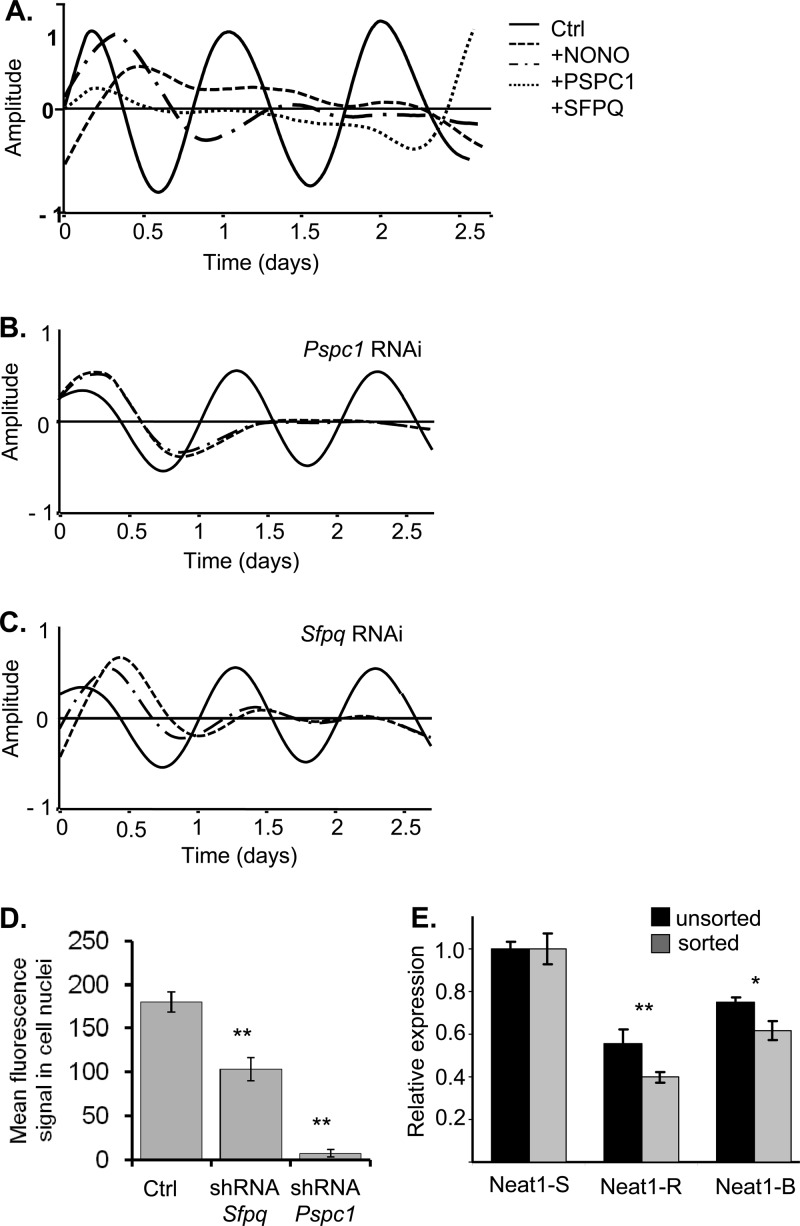
Fig 4.
(A) Bioluminescence from 3T3 cells transiently transfected with the Rev-Erbα-luc circadian reporter and constructs expressing either NONO, SFPQ, or PSPC1. Data shown are detrended and expressed in arbitrary units relative to mean expression. Ctrl, wild-type cells. Values for cells overexpressing NONO, PSPC1, and SFPQ are shown. (B and C) Bioluminescence from 3T3 cells transiently transfected with the Rev-Erbα-luc circadian reporter and RNAi constructs targeting either Pspc1 (B) or Sfpq (C). After synchronization with dexamethasone, cultures were measured 3 days. Data shown are detrended and expressed in arbitrary units relative to mean expression. Solid line, wild-type cells. Dashed lines, duplicate plates of cells expressing an Sfpq- or Pspc1-targeting vector. (D) Quantification of depletion of SFPQ and PSPC1 proteins from experiments above. Relative repression from 3T3 cells cotransfected with a green fluorescent protein-expressing plasmid and a plasmid expressing an RNAi interference construct targeting Sfpq or Pspc1. Averages shown are from 10 cells each (±standard errors). Mean fluorescence is expressed in arbitrary units. (E) Quantification (values are ±standard errors; n = 2 independent experiments, performed in triplicate) of Neat1 levels for two different RNAi constructs (R and B), as well as a scrambled hairpin (S) used in the experiment shown in Fig. 4, quantified from RNA of bulk-transfected cells (unsorted) or from cells cotransfected with a green fluorescent protein-expressing plasmid and then subjected to fluorescence-activated cell sorting to isolate green fluorescent protein-expressing cells (sorted).