Fig 2.
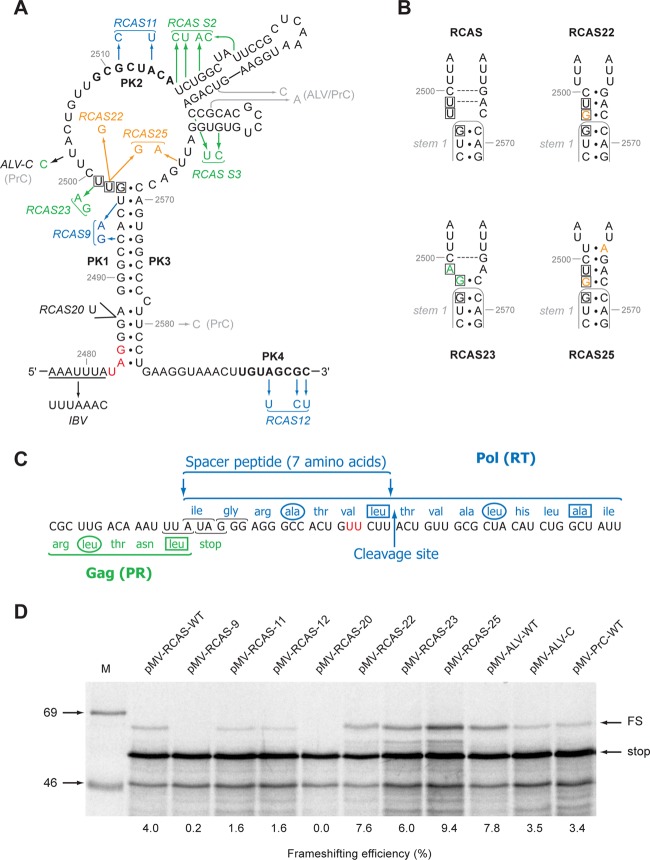
Mutational analysis of the RCAS frameshifting signal. (A) Mutations were introduced into pMV-RCAS by site-directed mutagenesis and are color coded to indicate the predicted effect on pseudoknot stability in orange (stabilization), blue (destabilization), or green (little effect/uncertain effect). Genomic sequence differences between RCAS and ALV/PrC are shown in gray. The valine codon (GUU) mutated in RCAS-22, -23, and -25 is boxed. The wild-type slippery sequence is underlined, and a variant with slippery sequence UUUAAAC is shown (IBV). The PK2 and PK4 regions, which form stem 2* of the pseudoknot, are in bold. The stop codon of the gag ORF is in red. (B) Predicted effect of the RCAS-23 and -25 mutations on the length of stem 1. The wild-type stem 1 is outlined in gray, and the mutated nucleotides are shown in the same colors as in panel A. Potential Watson-Crick base pairs are indicated by a dashed gray line. The valine codon, which in RCAS-22, -23, and -25 is mutated to a glycine codon, is boxed. (C) The protein sequences of Gag (green) and Pol (blue) in the overlap region are shown. The cleavage site at the C-terminal end of the seven-amino-acid spacer region is indicated. The two U residues mutated in RCAS-23 and RCAS-25 (see panel A), which change valine to glycine, are shown in red. Hydrophobic residues at position a (circled) and d (boxed) of a putative leucine zipper motif (45) (see the text) are indicated. (D) Ribosomal frameshift assays of pMV-RCAS variants. mRNAs derived from AflII-cut pMV-RCAS and derivatives were translated in RRL (at ∼50 μg/ml) for 1 hour, and the products resolved by 10% SDS-PAGE and visualized by autoradiography. Molecular size markers (in kDa) were also run on the gel (M). The 40-kDa nonframeshift (stop) and 54-kDa −1 frameshift products (FS) are indicated. The frameshifting efficiency measured for each signal is indicated below the relevant lanes and takes into account the number of methionines present in each product (stop, 11; FS, 11).