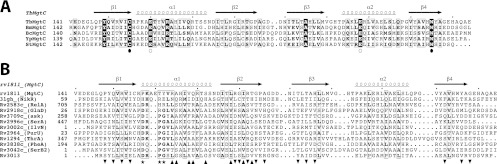
Fig 3.
Sequence alignment of MgtC proteins and ACT domains. (A) Sequence alignment of C-terminal domains of MgtC from pathogens which survive inside macrophages (M. tuberculosis, B. melitensis, B. cenocepacia, Y. pestis, and S. Typhimurium). Sequences were aligned manually, and the figure was drawn using the software program ESPript (11). Secondary structures were computed using the NMR structure of M. tuberculosis MgtC-Cter (this study). Four conserved residues that have been mutated to alanine during a previous analysis carried out with the S. Typhimurium MgtC protein (30) are indicated by circles. Black dots indicate residues that led to a loss of function when converted to alanine. (B) Sequence alignment of M. tuberculosis MgtC and the ACT domains identified in M. tuberculosis, as well as NikR from Helicobacter pylori. Mycobacterial proteins are identified by their gene names according to Tuberculist (http://genolist.pasteur.fr/TubercuList/). Sequences were aligned using the software program ViTO (6). We took advantage of the NMR structure of Rv1811 and the crystal structures available for Rv2996c (PDB 3DDN). For the other ACT domains, we modeled the 3D structure using close homologs detected with the server @TOME-2 (28) to help the refinement of our sequence-structure alignment. The figure was drawn using the program ESPript (11). The stars indicate the best-conserved residues that belong to the amino acid binding site of ACT domains. The positions buried by the dimerization of the ACT domain of NikR or of SerA (shown by up and down triangles, respectively) are highlighted in the alignment.