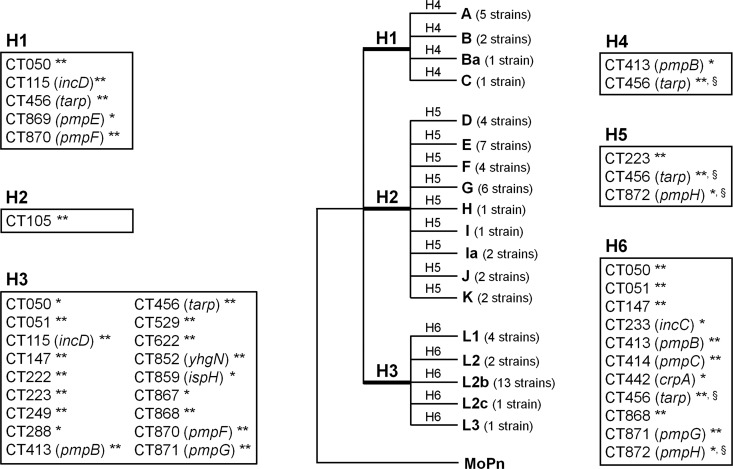
Fig 6.
Positive selection driving the directional C. trachomatis evolution toward niche-specific adaptation. The figure represents a model tree encompassing all 59 C. trachomatis strains that was created to facilitate a proper visualization of all biological hypotheses. These evaluate the existence of genes under positive selection (through the branch site test of positive selection) that may be involved in the following: specific cell appetence to columnar epithelial cells of ocular (H1) (serovars A to C) or genital (H2) (serovars D to K) mucosae and to mononuclear phagocytes (H3) (serovars L1 to L3) and pathogenic diversity among strains causing ocular disease (H4), genital disease (H5), or hemorrhagic proctitis and suppurative lymphadenitis (H6). This test was applied to each individual gene tree in an independent manner for each hypothesis, depending on the tree topology. The figure boxes show the genes found to be positively selected for each biological hypothesis. The likelihood-ratio test (LRT) was used to infer the statistical significance (P values) of positive selection in the foreground branches. ** and *, significance of P < 0.01 and P < 0.05, respectively; §, genes presenting congruent trees, but for which a specific biological hypothesis may be affected by recombination (49). See Table 1 for details on the positively selected codons.