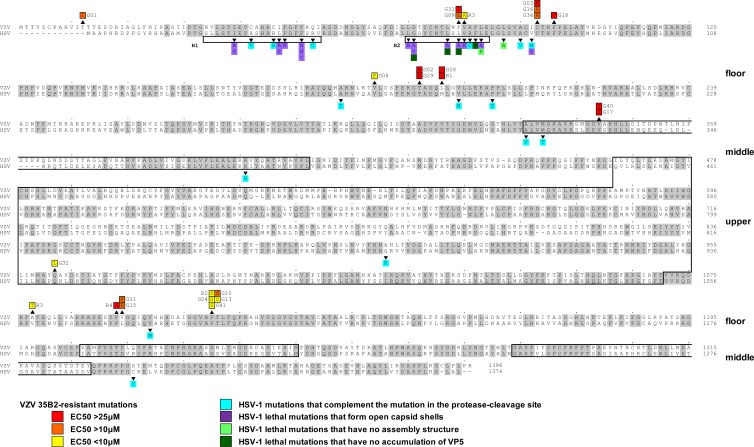
Fig 4.
Comparison of amino acid sequences between HSV-1 VP5 (UL19) and VZV MCP (ORF40) and locations of mutations identified in 35B2-resistant clones. Amino acid sequences identical in both HSV-1 VP5 and VZV MCP are highlighted in gray. Numbers at the ends of the sequences indicate the positions in the amino acid sequences. Mutations identified in 35B2-resistant clones are indicated above the VZV MCP sequences. The mutations in the clones for which EC50s were <10 μM, 10 to 25 μM, and >25 μM are highlighted in yellow, orange, and red, respectively. Shown beneath the HSV-1 VP5 sequence and highlighted in blue, purple, light green, and dark green, respectively, are the previously characterized HSV-1 VP5 mutations that complemented a virus defective in protease cleavage of the scaffold proteins (10, 58) and the lethal mutations that formed open capsid shells, those that formed no capsids, and those that did not accumulate VP5 in infected cells (20, 56). The HSV-1 VP5 helices, H1 and H2, and the previously predicted VP5 upper and middle domains (3, 4) are boxed.