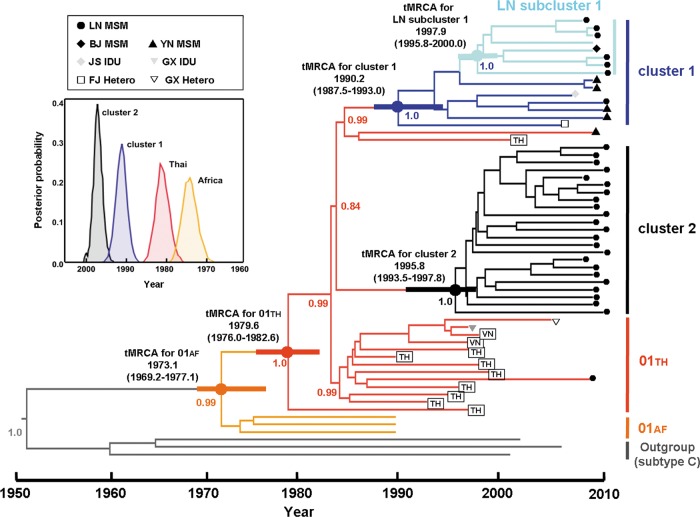
Fig 2.
Maximum clade credibility (MCC) tree representing the rooted genealogy of Liaoning CRF01_AE clusters. An MCC tree was obtained by Bayesian MCMC analysis based on nearly full-length HIV-1 nucleotide sequences (8,335 nt; HXB2, 803 to 9,181 nt) implemented in BEAST v 1.6.0. This particular tree was constructed based on a relaxed clock model in GTR+G4 combined with a constant coalescent model. HIV-1 subtype C sequences were used as outliers. The medians of tMRCAs and 95% credibility intervals (in parentheses) of CRF01_AE strains relevant to this study are as follows: African CRF01_AE ancestor, 1973.1 (1969.2 to 1977.1); Thailand CRF01_AE ancestor, 1979.6 (1976.0 to 1982.6); CRF01_AE cluster 1, 1990.2 (1987.5 to 1993.0); CRF01_AE cluster 2, 1995.8 (1993.5 to 1997.8); Liaoning (LN) subcluster 1, 1997.9 (1995.8 to 2000.0) (as determined using an GTR+γ4 relaxed clock model with a constant size model) (data not shown). The distribution of posterior probability of respective CRF01_AE clusters is illustrated in inset and shown as thick horizontal bars on the corresponding codes.