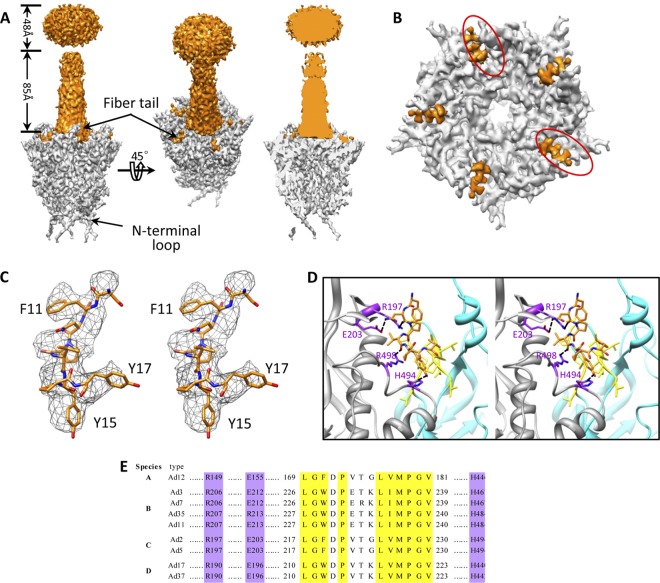
Fig 4.
Fiber-penton base interaction. (A) Density maps of a penton base (gray) and a fiber (orange) segmented from our 4.2-Å-resolution structure of the Ad5F35 virion (left and middle). Cut-away view of the density map of a penton base interacting with a fiber shows the absence of densities inside the central pore (right). (B) Penton base (gray) and fiber tail (orange). The fiber has been computationally removed for clarity. Five fiber tail densities are visible due to the imposed 5-fold symmetry; two positions (circled) should be left unoccupied. (C) Cross-eye stereo view of the atomic model of the fiber tail (amino acids 11 to 17) superimposed on its density map (mesh). (D) Cross-eye stereo view of the interactions (ribbon) between the fiber tail (orange stick view) and the two copies of adjacent penton base proteins (gray and cyan ribbons) show a hydrophobic groove formed by a number of hydrophobic residues (yellow) in one subunit of the penton base protein (cyan) and a few putative hydrogen bonds (purple) involving residues Asn12 (fiber) and Glu203, Asn12 (fiber) and Arg197, Val14 (fiber) and Arg496, and Tyr15 (fiber) and His494 between the fiber tail and another subunit of the penton base protein (gray). (E) Amino acid sequence alignment of penton base protein from different human Ad serotypes in six species (A to F) shows that the residues involved in hydrophobic interactions (yellow) and hydrogen interactions (purple) with fiber tail are fully conserved.