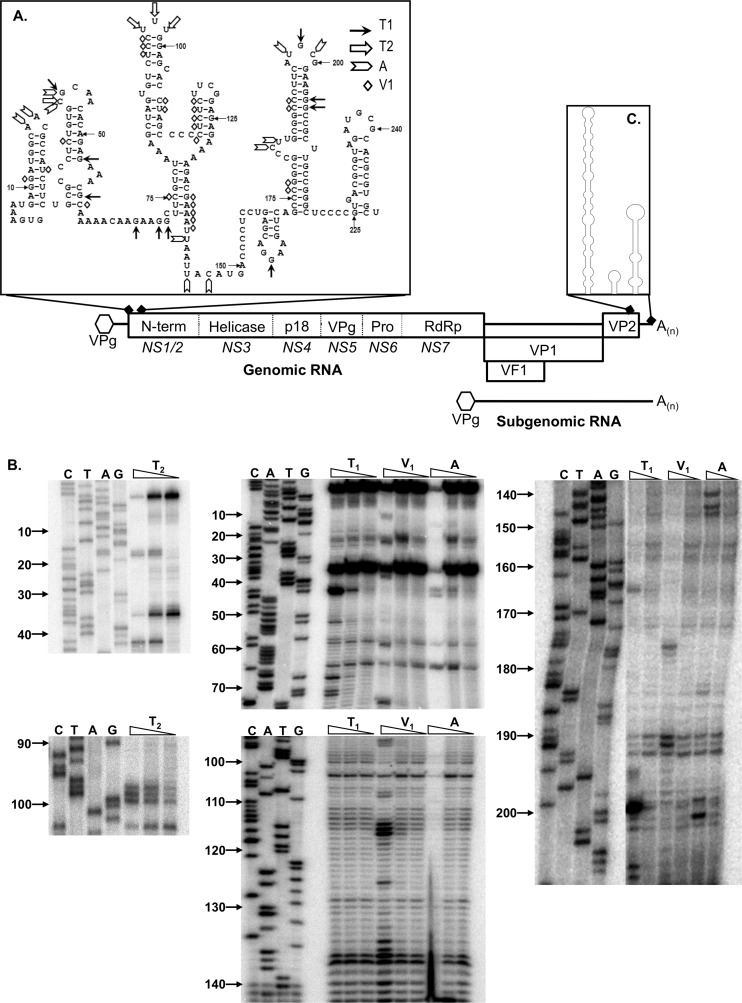
Fig 1.
Murine norovirus genome organization and positions of conserved RNA structures. The NS1 to -7 nomenclature, as proposed by Sosnovtsev et al. (66), is presented in italics below the various open reading frames. Note that the RNA structures are not drawn to scale. (A) Predicted RNA structures adopted by the 5′ end of the MNV genome, highlighting the biochemically determined cleavage sites for single-stranded RNA-specific RNases T1, T2, and A, along with the double-stranded RNA-specific RNase V1 cleavage sites. (B) Primary data for RNase sensitivity mapping of the 5′ end of the MNV genome. In vitro-transcribed RNA encompassing the region was subjected to limited RNase digestion and subsequent primer extension analysis as described previously. A sequencing ladder prepared using the same primer was also performed to allow the identification of the RNase cleavage sites. Analysis was performed a minimum of 3 times, and 1 representative gel shown. Nucleotide positions are numbered according to their positions in the murine norovirus genome. Note that data are shown for regions containing RNase cleavage sites only. (C) Schematic illustration of the RNA structures adopted by the 3′ 237 nucleotides of the MNV genome. For clarity, the sequence of this region is not shown. The structure shown is as previously computationally predicted and biochemically confirmed (3).