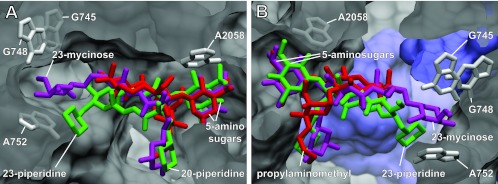
Fig 2.
The macrolide site in the ribosome tunnel. (A and B) Two views of superimposed binding sites for tildipirosin (green), tilmicosin (magenta), and tulathromycin (red). Parameters for calculating tildipirosin and tilmicosin (and tylosin) binding using Schrödinger 2010(U1) Suite software were described recently (18). The model presented here for tulathromycin on the E. coli ribosome was calculated in a similar manner based on the Thermus thermophilus ribosome-azithromycin cocrystal structure PDB 3OI1 (1) aligned against the E. coli 50S structure PDB 3OAT (5). Clear overlap in the positions of the 5-amino sugars mycaminose (tildipirosin and tilmicosin) and desosamine (tulathromycin) is evident. The 20-piperidine (tildipirosin), the 20-dimethylpiperidine (tilmicosin), and the 3-cladinose propylaminomethyl side chain (tulathromycin) reach into the tunnel lumen with slightly different orientations. More distinct differences are seen in the locations of the 23-piperidine (tildipirosin) and the 23-mycinose (tilmicosin), while tulathromycin has no equivalent structure. Ribosomal nucleotides interacting with the drug substituents are indicated as gray sticks; ribosomal proteins are shown in blue.