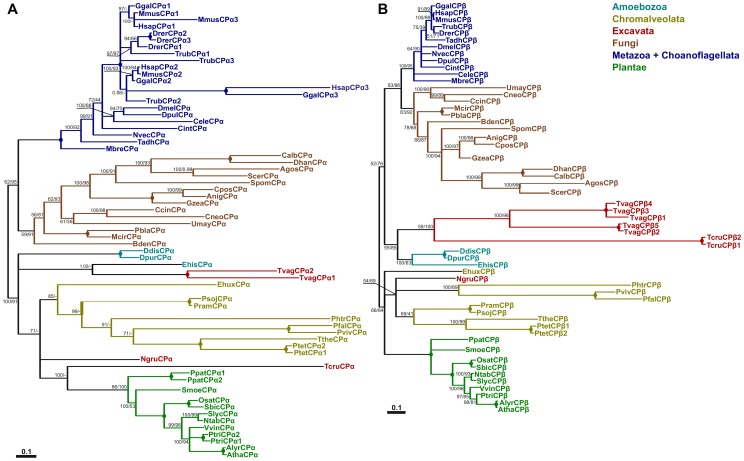
Figure 1. Phylogenetic analysis of CPα (A) and CPβ (B).
Both trees represent protein bayesian phylogeny of particular genes. Numbers at nodes correspond to posterior probabilities from Bayesian analysis and the approximate likelihood ratio test with SH-like (Shimodaira-Hasegawa-like) support from maximum likelihood method, respectively. Circles represent support 100% by both methods, Missing values indicate support below 50%, dash indicates that a different topology was inferred by ML method. Branches were collapsed if inferred topology was not supported by both methods. Scale bar indicating the rates of substitutions/site is shown in corresponding tree. Abbreviations used: Agos – Ashbya gossypii, Alyr – Arabidopsis lyrata, Anig – Aspergillus niger Atha – Arabidopsis thaliana, Bden – Batrachochytrium dendrobatidis, Calb – Candida albicans, Ccin – Coprinopsis cinerea, Cele – Caenorhabditis elegans, Cint – Ciona intestinalis, Cneo – Cryptococcus neoformans, Cpos – Coccidioides posadasii, Ddis – Dictyostelium discoideum, Dhan – Debaryomyces hansenii, Dmel – Drosophila melanogaster, Dpur – Dictyostelium purpureum, Dpul – Daphnia pulex, Drer - Danio rerio, Ehis - Entamoeba histolytica, Ehux - Emiliania huxleyi, Ggal – Gallus gallus, Gzea – Gibberella zeae, Hsap – Homo sapiens, Mbre – Monosiga brevicollis, Mcir – Mucor circinelloides, Mmus – Mus musculus, Ngru – Naegleria gruberi, Ntab – Nicotiana tabacum, Nvec – Nematostella vectensis, Osat – Oryza sativa, Pbla – Phycomyces blakesleeanus, Pfal – Plasmodium falciparum, Phtr – Phaeodactylum tricornutum, Ppat – Physcomitrella patens, Pram – Phytophthora ramorum, Psoj – Phytophthora sojae, Ptet - Paramecium tetraurelia, Ptri – Populus trichocarpa, Pviv – Plasmodium vivax, Sbic – Sorghum bicolor, Scer – Saccharomyces cerevisiae, Slyc – Solanum lycopersicum, Spom – Schizosaccharomyces pombe, Smoe – Selaginella moellendorffi, Tadh – Trichoplax adherans, Tcru – Trypanosoma cruzi, Trub – Takifugu rubripes, Tthe – Tetrahymena thermophila, Tvag – Trichomonas vaginalis, Umay – Ustilago_maydis and Vvin – Vitis vinifera.