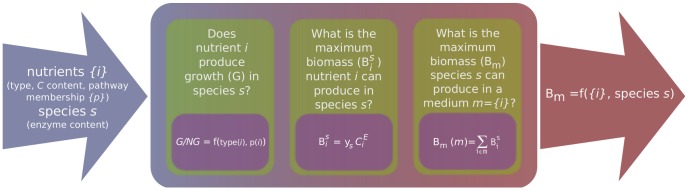
Figure 1. Schematic representation of the development of a model for maximum biomass production in complex media of microbial organisms.
We aim at developing a phenomenological model to predict the maximum biomass production B
m of species s when growing in a medium containing a set of nutrients {i} acting as a carbon source under aerobic conditions. That is, we want to express B
m as a function f ({i}, s) that only takes into account data related to: i) the set of nutrients {i} available, namely, nutrient type, the set of pathways {p(i)} that can catabolize each nutrient, and the carbon content Ci of each nutrient; and ii) the species s, specifically the presence or not of certain enzymes in a species that allow to catabolize specific types of nutrients (enzymes EC: 1.1.1.35, EC: 2.3.1.16, and EC 3.5.2.17 - see text). In order to achieve our goal, there are three different questions we need to answer: i) Does nutrient i produce growth or not in species s when acting as the sole source of carbon? We find that whether nutrient i produces growth (G) or not (NG) is a function of the nutrient type (see text) and its pathway membership; (ii) If a nutrient produces growth, what is the maximal biomass 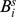 it can produce in species s when acting as the sole source of carbon? We find that
it can produce in species s when acting as the sole source of carbon? We find that 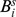 is proportional to Ci, the number of carbons in nutrient i, and that the proportionality constant y
s depends on the species s. iii) What is the maximal biomass production B
m(m) when growing on a complex medium m? We find that B
m(m) can be well approximated by adding up the individual contributions
is proportional to Ci, the number of carbons in nutrient i, and that the proportionality constant y
s depends on the species s. iii) What is the maximal biomass production B
m(m) when growing on a complex medium m? We find that B
m(m) can be well approximated by adding up the individual contributions 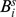 of nutrients i present in medium m.
of nutrients i present in medium m.