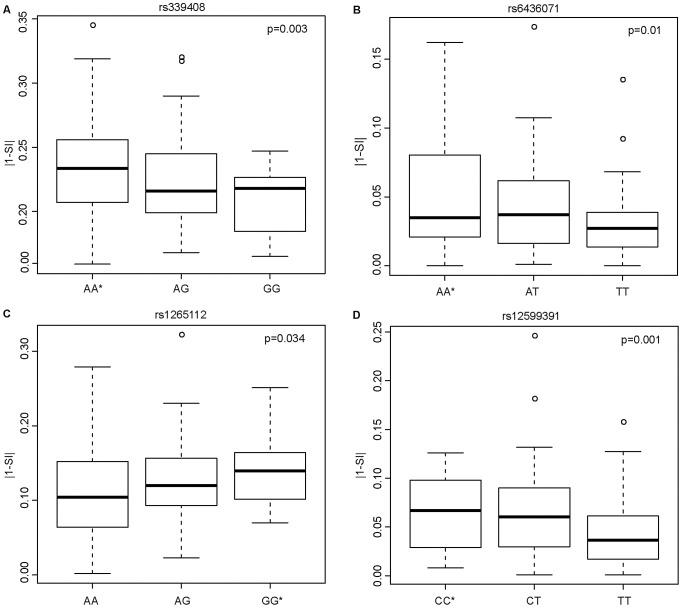
Figure 4. Candidate ISE SNPs Associated with an Increase in Exon Skipping.
The y-axis is the absolute value of 1 minus SI (|1-SI|); here, a higher level of |1-SI| corresponds to a higher level of exon skipping. The allele with an asterisk showed an association with increased exon skipping. In the case of the SNP rs339408 (with alleles A/G), the G allele is located within three ISE motifs, namely at the second, third, and fourth site of AGGGAT, CAGGGA, and TCAGGG, respectively, but the A allele is the exon skipping associated ISE allele (A). In the case of the SNP rs6436071 (A/T), the T allele participates in two ISE motifs, namely at the second and third site of CTTGGC and GCTTGG, respectively (B). In the case of rs1265112 (A/G), the A allele is located at the fourth site in the CACACT ISE motif but the G allele is the exon skipping associated ISE allele (C). Finally, for rs12599391 (C/T), the T allele is located at the 3rd site of the AATTGT ISE motif but the C allele is the exon skipping associated ISE allele (D).