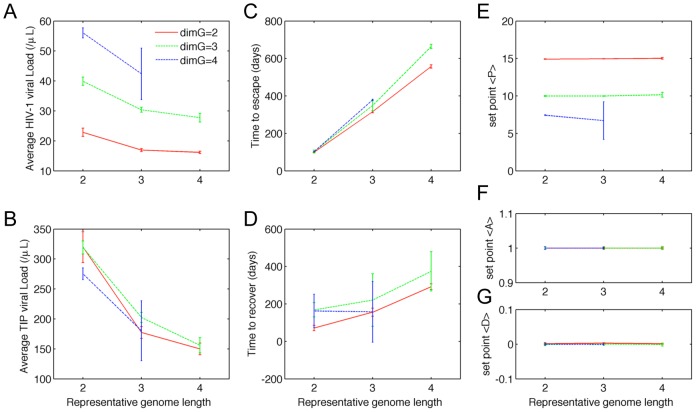
Figure 4. The co-evolutionary dynamics are qualitatively robust to variation in genome size and dimension.
We tested model sensitivity to the dimension of genome space (i.e. the number of values each genome site could have; denoted dimG) and the genome length (i.e. the number of sites corresponding to each parameter. The co-evolutionary dynamics of HIV-1 and TIP were characterized by 7 attributes: the mean HIV-1 and TIP viral loads (panel (A) and (B)) over the whole simulation (3000 days), the lengths of the PR phase and the ES phase (panel (C) and (D)), and the time-average mean values of P, A and D of the population of dually infected cells (panel (E), (F) and (G)) during the ‘set-point’ phase. Data points and error bars correspond to the mean and standard deviation of 100 realizations of the model for those parameter values.