Figure 1.
Identification of the Disease-Causing PNPT1 Mutation in a Consanguineous Family Affected by Nonsyndromic Hearing Loss
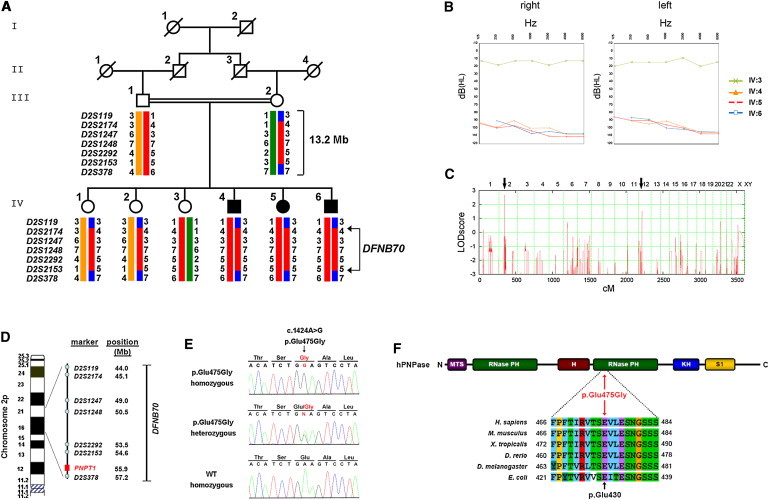
(A) Pedigree of the DFNB70-linked family. Microsatellite haplotypes indicate homozygosity between markers D2S119 and D2S378 in affected family members (filled symbols). According to the Genome Reference Consortium release h37, February 2009, the linked region spans approximately 13.2 Mb.
(B) Pure-tone audiograms of the three affected children with severe hearing loss; an audiogram of an unaffected sibling is shown for comparison. The following abbreviations are used: dB(HL), decibel hearing loss; and Hz, frequency in Hertz.
(C) Genome-wide homozygosity mapping identified two putative loci (black arrows) with multipoint LOD scores of 2.7 and 1.5 on chromosomes 2 and 12, respectively (see also Figure S1).
(D) Illustration shows the position of the DFNB70 locus on the short arm of chromosome 2; PNPT1 is at the centromeric end (see also Figure S1 and Table S1).
(E) Direct sequencing identified a homozygous c.1424A>G mutation in PNPT1 in affected individuals (upper electropherogram) and in a heterozygous state in unaffected carriers (middle). A homozygous wild-type (WT) sequence is shown on the bottom. Amino acid abbreviations are shown above the sequence traces.
(F) A PNPase monomer consists of two RNase-PH domains, which are separated by an α-helix, and two RNA binding domains, KH and S1. Additionally, mammalian PNPase carries an N-terminal mitochondrial targeting sequence (MTS). The multiple-sequence alignment was generated with the ClustalW2 Multiple Sequence Alignment tool and depicts conservation of the crucial glutamic acid residue. Amino acid position 430 in E. coli corresponds to position 475 in humans.
