Figure 5.
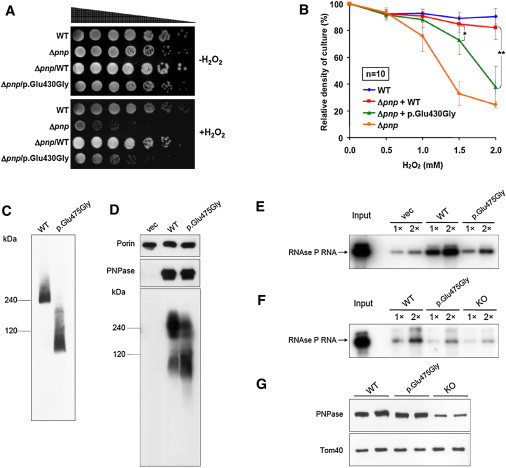
Functional Consequences of the Identified PNPT1 Mutation
(A–B) Complementation experiments in bacteria: E. coli pnp-knockout strain (Δpnp) was transformed with constructs expressing either bacterial WT or mutant (p.Glu430Gly) pnp. E. coli K-12 strain BW25113 was used as a WT reference.
(A) Spot tests. Strains were serially diluted and grown on lysogeny broth (LB) agar plates either with or without 0.6 mM hydrogen peroxide. Growth of Δpnp is suppressed by hydrogen peroxide and can be almost completely restored by WT Pnp but only partially by p.Glu430Gly Pnp.
(B) Indicated strains were grown in LB medium and exposed to increasing concentrations of hydrogen peroxide. Exogenous expression of WT pnp restores growth significantly better than does expression of the pnp mutant encoding p.Glu430Gly (Student’s two-tailed t test: ∗p = 7.06 × 10−3, ∗∗p = 2.56 × 10−6). Error bars indicate standard deviation of ten independent experiments.
(C–D) The assembly state of exogenous WT and p.Glu475Gly PNPase was determined by nondenaturing blue-native gel electrophoresis.
(C) In MEFs with floxed endogenous mouse Pnpt1 knocked down by CMV-CRE recombinase-induced partial knockout, the trimerization of exogenous p.Glu475Gly PNPase is severely impaired and shifted toward monomers.
(D) In the GA74-1A yeast strain, p.Glu475Gly PNPase colocalized with a mitochondrial marker, PORIN, and showed a protein level that was similar to that of exogenous WT PNPase. However, the assembly of p.Glu475Gly PNPase in a higher-order, trimeric PNPase complex was impaired compared to WT PNPase, as shown by a significant reduction of the trimer band at ∼240 kDa and increase of the monomer band at ∼85 kDa.
(E) RNA import assay in yeast. In-vitro-transcribed human RNase P RNA was incubated with yeast mitochondria expressing WT PNPT1, a PNPT1 mutant coding for p.Glu475Gly, or an empty vector (“vec”). Import reactions were repeated with 1× and 2× amounts of RNA. Abundance of imported RNase P RNA in mitochondria containing p.Glu475Gly PNPase is one quarter of the level of that in mitochondria containing WT PNPase.
(F) RNA-import assay in MEFs. In-vitro-transcribed human RNase P RNA was incubated with mitochondria isolated from different MEF lines: WT (mouse Pnpt1 partial knockout plus exogenous WT PNPT1 expression), p.Glu475Gly (Pnpt1 partial knockout plus exogenous PNPT1 expression encoding p.Glu475Gly), or knockout (Pnpt1 expression reduced by 75%). Import reactions were repeated with 1× and 2× amounts of RNA. Import of RNase P RNA is two times lower in PNPT1-mutant-expressing mitochondria than in WT-PNPT1-transduced mitochondria.
(G) Immunoblot of PNPase and mitochondria-localized TOM40 from different MEF lines.
