Figure 1.
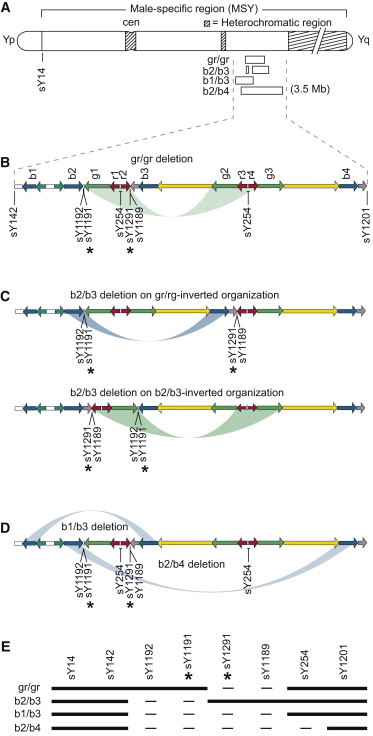
The AZFc Region of the Y Chromosome and Deletions Affecting It
(A) Overview of the Y chromosome, including the male-specific region (MSY). The AZFc region is located in the euchromatic portion of the long arm. STS sY14, located in the sex-determining gene SRY (MIM 480000), served as a positive control for the presence of detectable Y chromosome DNA. The four deletions observed in the present study are schematized here and in the panels below.
(B) Sequence organization of the AZFc region and STSs used for detecting and categorizing deletions involving AZFc. Colored arrows indicate large, nearly identical segmental duplications, termed “amplicons” in this context. Arrows of the same color are >99.82% identical to each other.1Table S1 provides assay details for STSs. We used STSs sY1191 and sY1291, marked by asterisks, in stage 1 of the screen. STS sY254 detects multiple sites in the AZFc region. A green “bow” indicates amplicons involved in ectopic crossing over in the gr/gr deletion and regions and STSs affected; the gr/gr deletion results in loss of only sY1291 and sY1189.
(C) The b2/b3 deletion can arise on the two inverted variants of the AZFc region shown, but not on the reference organization;2 the b2/b3 deletion arising on either of the two inverted variants results in the same organization of amplicons and results in the loss of only sY1192 and sY1191.
(D) Ectopic crossing over and the extents of the b1/b3 and b2/b4 deletions. The b1/b3 deletion (upper bow) results in loss of all copies of STSs sY1192, sY1191, sY1291, and sY1189 but spares two copies of sY254, as well as sY142 and sY1201. The b2/b4 deletion (lower bow) has a similar pattern but removes all copies of sY254.
(E) Patterns of PCR positives and negatives used for identifying four types of recurrent deletion. Black indicates the presence of STS PCR product, and “−” indicates absence.
