Figure 2.
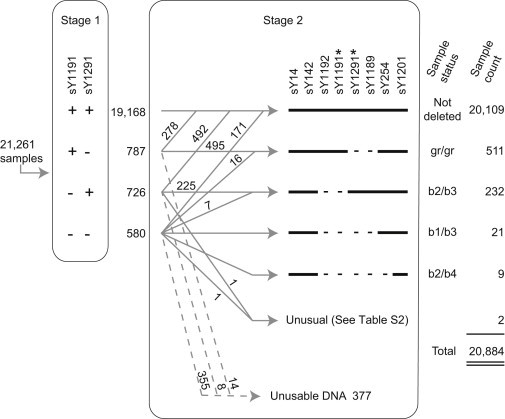
Workflow for Detecting and Categorizing Deletions Involving AZFc
In stage 1, we tested 21,261 DNA samples for the presence of STSs sY1191 and sY1291; 19,168 samples were scored positive for both STSs. The 2,093 samples that were scored negative for one or both STSs were subject to further testing in stage 2 with the use of the STSs shown. Also, sY1191 and sY1291, marked by asterisks, were retested. This allowed us to categorize the deletions as shown. As expected, stage 2 retesting revealed that some STSs deemed absent in stage 1 were in fact present. For example, 492 samples that were scored negative for sY1191 in stage 1 were scored positive in stage 2. This was because we set a liberal threshold in stage 1 for scoring sY1191 absence to avoid missing deletions. We also determined in stage 2 that 377 DNA samples were not usable; we could not reliably amplify positive control STSs in these samples. Thus, the total number of samples assayed was 20,884.
