Abstract
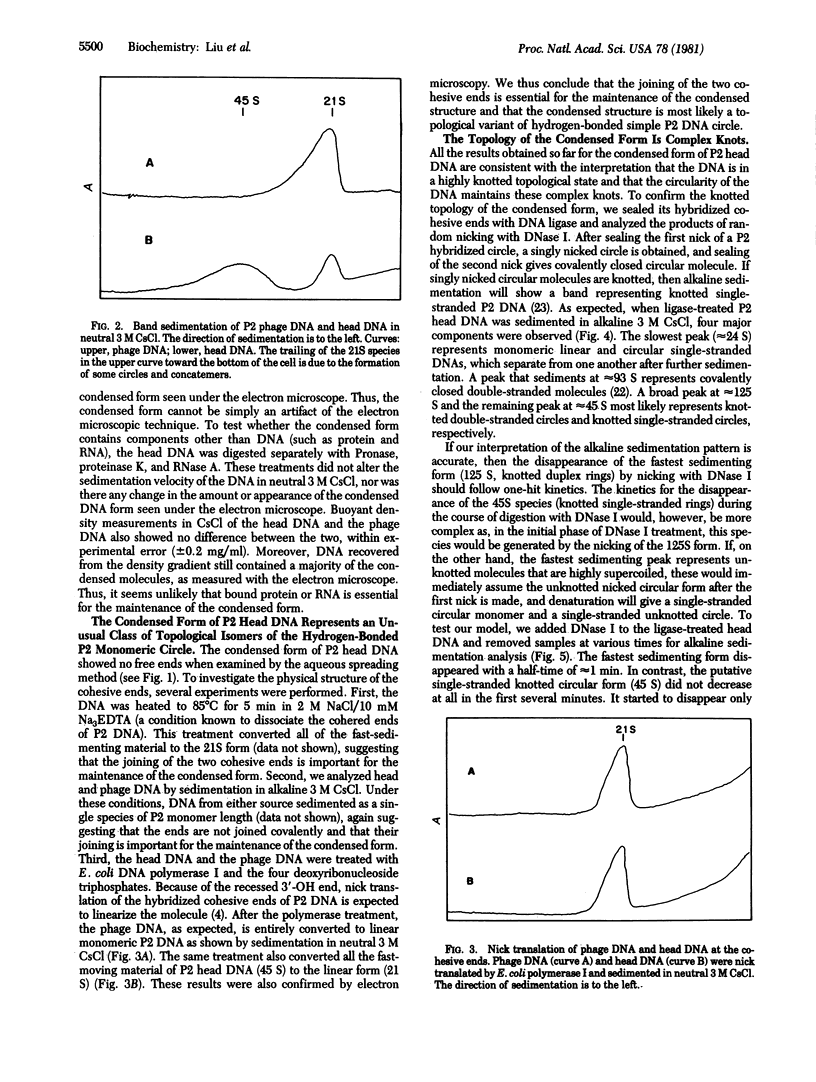
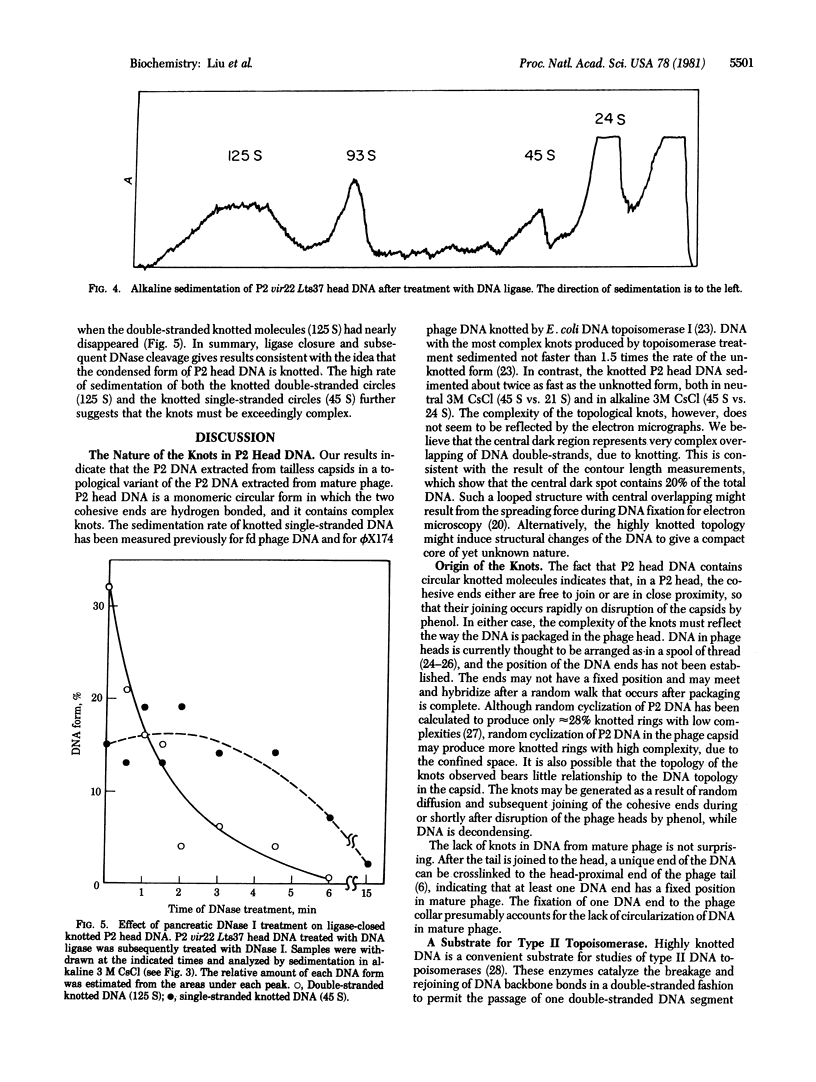
The majority of the DNA prepared from tailless capsids of bacteriophage P2 by the phenol extraction procedure consists of monomeric rings that have their cohesive ends joined. Electron microscopic and ultracentrifugal studies indicate that these molecules have a complex structure that is topologically knotted; they have a more compact appearance and a higher sedimentation coefficient when compared with regular nicked P2 DNA rings. Linearization of these rings by thermal dissociation or repair of the cohesive ends by DNA polymerase I in the presence of all four deoxynucleoside triphosphates gives molecules that are indistinguishable from normal P2 DNA that has been similarly treated. The knotted nature of the majority of P2 head DNA is further supported by analyzing the products when these molecules are treated with ligase and the ligase-treated molecules are subsequently nicked randomly with DNase I. The data are consistent with the notion that, if such a molecule is first converted to a form that contains only one single-chain scission per molecule, strand separation gives a linear strand and a highly knotted single-stranded ring. The results suggest that the DNA packaged in tailless P2 capsids is arranged in a way that leads to the formation of a complex knot when the ends join. In an intact phage particle, the anchoring of one terminus of the DNA to the head-proximal end of the tail [Chattoraj, D. K. & Inman, R. B. (1974) J. Mol. Biol. 87, 11-22] presumably diminishes or prevents this kind of joining. The novel knotted DNA can be used to assay type II DNA topoisomerases that break and rejoin DNA in a double-stranded fashion.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERTANI L. E. The effect of the inhibition of protein synthesis on the establishment of lysogeny. Virology. 1957 Aug;4(1):53–71. doi: 10.1016/0042-6822(57)90043-0. [DOI] [PubMed] [Google Scholar]
- Bertani G. Deletions in bacteriophage P2. Circularity of the genetic map and its orientation relative to the DNA denaturation map. Mol Gen Genet. 1975;136(2):107–137. doi: 10.1007/BF00272034. [DOI] [PubMed] [Google Scholar]
- Bertani G., Ljungquist E., Jagusztyn-Krynicka K., Jupp S. Defective particle assembly in wild type P2 bacteriophage and its correction by the lg mutation. J Gen Virol. 1978 Feb;38(2):251–261. doi: 10.1099/0022-1317-38-2-251. [DOI] [PubMed] [Google Scholar]
- Brown P. O., Cozzarelli N. R. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979 Nov 30;206(4422):1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- Chattoraj D. K., Inman R. B. Location of DNA ends in P2, 186, P4 and lambda bacteriophage heads. J Mol Biol. 1974 Jul 25;87(1):11–22. doi: 10.1016/0022-2836(74)90556-7. [DOI] [PubMed] [Google Scholar]
- Chattoraj D. K., Inman R. B. Position of two deletion mutations on the physical map of bacteriophage P2. J Mol Biol. 1972 May 28;66(3):423–434. doi: 10.1016/0022-2836(72)90424-x. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Casjens S. R. DNA packaging by the double-stranded DNA bacteriophages. Cell. 1980 Sep;21(2):319–331. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Harrison S. C. DNA arrangement in isometric phage heads. Nature. 1977 Aug 18;268(5621):598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetskii M. D., Lukashin A. V., Vologodskii A. V. Statistical mechanics and topology of polymer chains. Nature. 1975 Dec 4;258(5534):398–402. doi: 10.1038/258398a0. [DOI] [PubMed] [Google Scholar]
- Inman R. B., Bertani G. Heat denaturation of P2 bacteriophage DNA: compositional heterogeneity. J Mol Biol. 1969 Sep 28;44(3):533–549. doi: 10.1016/0022-2836(69)90378-7. [DOI] [PubMed] [Google Scholar]
- Inman R. B., Schnös M., Simon L. D., Six E. W., Walker D. H., Jr Some morphological properties of P4 bacteriophage and P4 DNA. Virology. 1971 Apr;44(1):67–72. doi: 10.1016/0042-6822(71)90153-x. [DOI] [PubMed] [Google Scholar]
- Jovin T. M., Englund P. T., Bertsch L. L. Enzymatic synthesis of deoxyribonucleic acid. XXVI. Physical and chemical studies of a homogeneous deoxyribonucleic acid polymerase. J Biol Chem. 1969 Jun 10;244(11):2996–3008. [PubMed] [Google Scholar]
- Koller T., Sogo J. M., Bujard H. An electron microscopic method for studying nucleic acid-protein complexes. Visualization of RNA polymerase bound to the DNA of bacteriophages T7 and T3. Biopolymers. 1974 May;13(5):995–1009. doi: 10.1002/bip.1974.360130514. [DOI] [PubMed] [Google Scholar]
- Lengyel J. A., Goldstein R. N., Marsh M., Sunshine M. G., Calendar R. Bacteriophage P2 head morphogenesis: cleavage of the major capsid protein. Virology. 1973 May;53(1):1–23. doi: 10.1016/0042-6822(73)90461-3. [DOI] [PubMed] [Google Scholar]
- Lindahl G. On the control of transcription in bacteriophage P2. Virology. 1971 Dec;46(3):620–633. doi: 10.1016/0042-6822(71)90065-1. [DOI] [PubMed] [Google Scholar]
- Lindqvist B. H. Vegetative DNA of temperate coliphage P2. Mol Gen Genet. 1971;110(2):178–196. doi: 10.1007/BF00332647. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Depew R. E., Wang J. C. Knotted single-stranded DNA rings: a novel topological isomer of circular single-stranded DNA formed by treatment with Escherichia coli omega protein. J Mol Biol. 1976 Sep 15;106(2):439–452. doi: 10.1016/0022-2836(76)90095-4. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Liu C. C., Alberts B. M. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980 Mar;19(3):697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Miller K. G. Eukaryotic DNA topoisomerases: two forms of type I DNA topoisomerases from HeLa cell nuclei. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3487–3491. doi: 10.1073/pnas.78.6.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M. Infectivity of phage P2 DNA in presence of helper phage. Mol Gen Genet. 1967;99(1):88–96. doi: 10.1007/BF00306461. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Fisher L. M., O'Dea M. H., Gellert M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P., Anraku Y., Lehman I. R. Deoxyribonucleic acid ligase. Isolation and physical characterization of the homogeneous enzyme from Escherichia coli. J Biol Chem. 1973 Nov 10;248(21):7495–7501. [PubMed] [Google Scholar]
- Murray K., Murray N. E. Terminal nucleotide sequences of DNA from temperate coliphages. Nat New Biol. 1973 May 30;243(126):134–139. doi: 10.1038/newbio243134a0. [DOI] [PubMed] [Google Scholar]
- Padmanabhan R., Wu R., Calendar R. Complete nucleotide sequence of the cohesive ends of bacteriophage P2 deoxyribonucleic acid. J Biol Chem. 1974 Oct 10;249(19):6197–6207. [PubMed] [Google Scholar]
- Pruss G. J., Calendar R. Maturation of bacteriophage P2 DNA. Virology. 1978 May 15;86(2):454–467. doi: 10.1016/0042-6822(78)90085-5. [DOI] [PubMed] [Google Scholar]
- Richards K. E., Williams R. C., Calendar R. Mode of DNA packing within bacteriophage heads. J Mol Biol. 1973 Aug 5;78(2):255–259. doi: 10.1016/0022-2836(73)90114-9. [DOI] [PubMed] [Google Scholar]
- Sasaki I., Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Microbiol. 1965 Sep;40(3):365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- Sunshine M. G., Thorn M., Gibbs W., Calendar R., Kelly B. P2 phage amber mutants: characterization by use of a polarity suppressor. Virology. 1971 Dec;46(3):691–702. doi: 10.1016/0042-6822(71)90071-7. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Martin K. V., Calendar R. On the sequence similarity of the cohesive ends of coliphage P4, P2, and 186 deoxyribonucleic acid. Biochemistry. 1973 May 22;12(11):2119–2123. doi: 10.1021/bi00735a016. [DOI] [PubMed] [Google Scholar]



