Table 1. Time (seconds) needed to complete one update of all coordinates and to reach convergence in sets of gene expression data from blood disorders.
| rNMF | control | ||||
| Data set, reference | Data size | iteration | convergence | iteration | convergence |
| Acute Myeloid Leukemia [36] | 22283×293 | 0.75 | 21.7 | 2.2 | 219.4 |
| Acute Myeloid Leukemia [37] | 54613×461 | 3.95 | 128.8 | 10.2 | >600 |
| Acute Myeloid Leukemia [38] | 44692×162 | 0.96 | 17.3 | 1.5 | 163.6 |
| Acute Lymphoblastic Leukemia [39] | 22215×288 | 0.94 | 17.8 | 2.3 | 245.7 |
| Multiple Myeloma [40] | 54613×320 | 3.04 | 29.1 | 6.4 | >600 |
All methods were implemented in C++ and identically initialized. Timings obtained on a 2.30 GHz Intel Core i7 2820QM CPU with 16 GB RAM. For convergence, we required a relative decrease in the objective function less than 10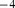 in successive iterations. Throughout,
in successive iterations. Throughout, 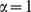 and
and 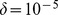 .
.
