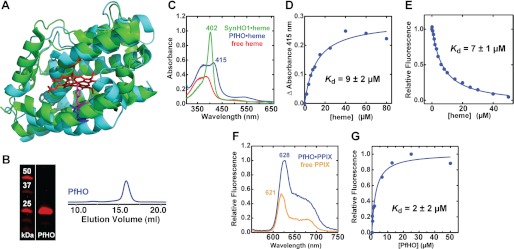
FIGURE 6.
Heme binding by PfHO versus SynHO1. A, structural alignment of the x-ray crystallographic model of SynHO1 containing bound heme (green, Protein Data Bank code 1WE1) with the Rosetta-derived model of the HO domain of PfHO (cyan) (root mean square deviation = 1.4 Å). The bound heme is shown in red; the distal Gly residue and the proximal His ligand of SynHO1 are shown in orange and violet, respectively, and the aligned Lys residue in PfHO is shown in blue. B, Coomassie-stained SDS-polyacrylamide gel and gel filtration chromatogram of purified PfHO. C, UV-visible absorbance spectrum of 10 μm heme free in solution (red) or bound to 20 μm SynHO1 (green) or 200 μm PfHO (blue). D, heme binding to 2 μm PfHO leads to a saturable absorbance increase at 415 nm. Plotting this increase as a function of heme concentration and fitting with a quadratic binding equation (R2 = 0.98) gives an apparent dissociation constant (Kd) of 9 ± 2 μm. E, heme binding to PfHO leads to quenching of endogenous Trp fluorescence at 387 nm. Fitting this fluorescence decrease with a quadratic binding equation (R2 = 0.99) gives an apparent affinity of 7 ± 1 μm. F, fluorescence emission spectrum (excitation 400 nm) of 10 μm PPIX free in solution (orange) or bound to 20 μm PfHO (blue). G, binding of 0.1 μm PPIX to PfHO leads to a saturable increase in PPIX fluorescence intensity. Plotting the fluorescence increase at 628 nm versus PfHO concentration and fitting with a quadratic binding equation (R2 = 0.97) yields an apparent Kd of 2 ± 2 μm.