FIGURE 9.
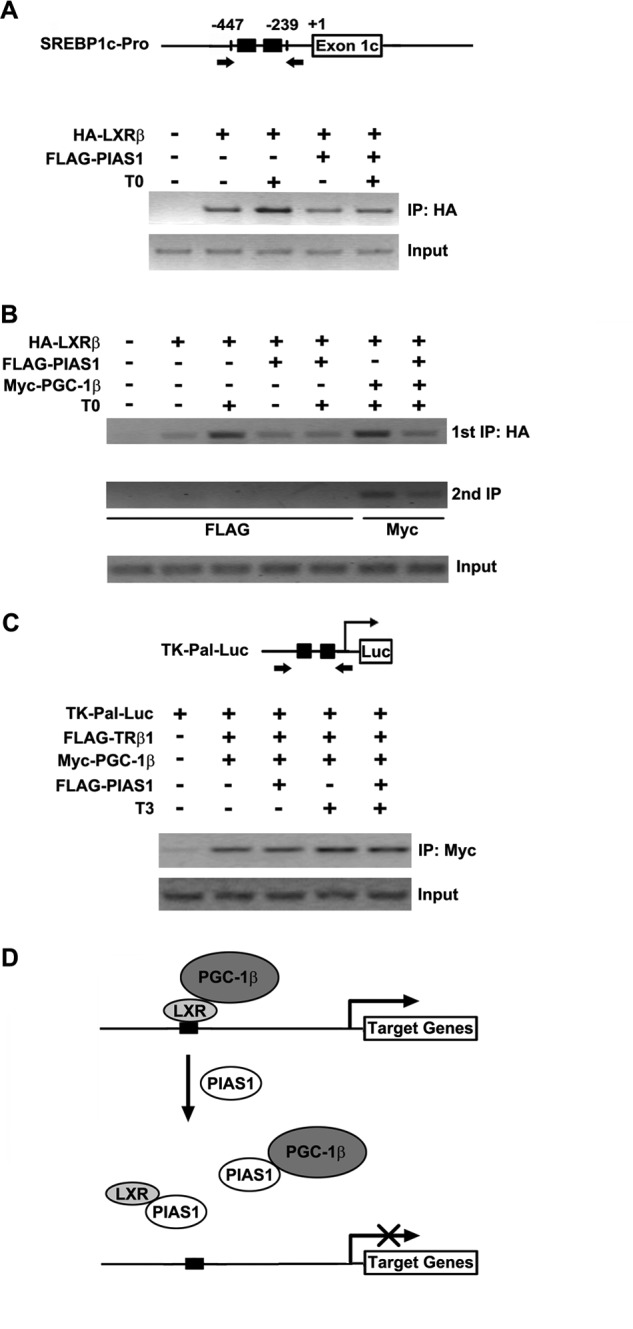
PIAS1 impairs the ability of LXRβ to bind to its target DNA sequence. A, schematic for the Srebp1c promoter containing the indicated LXREs. The arrows flanking the LXREs indicate the primers used for the ChIP assays in A and B. 293T cells were transfected for 48 h with empty vector (−) or the indicated plasmids and then treated with DMSO or 5 μm T0901317 for 12 h. Chromatin extracts were immunoprecipitated (IP) with anti-HA antibody, and the precipitated genomic DNA was subjected to PCR amplification. Cellular genomic DNA was used as the input control. B, 293T cells were co-transfected for 48 h with empty vector (−) or the indicated plasmids. Cells were likewise treated prior to chromatin immunoprecipitation with anti-HA antibody. The immunoprecipitates were then used for a secondary immunoprecipitation with anti-FLAG or anti-Myc antibody as indicated. The precipitated DNA was subjected to PCR amplification with cellular genomic DNA used as the input control. Representative results are shown from at least two independent experiments. C, schematic for the TK-Pal-Luc construct containing the indicated TREs. The arrows flanking the TREs indicate the primers used for the ChIP assay. 293T cells were transfected for 48 h with empty vector (−) or the indicated plasmids and then treated with DMSO or 100 nm T3 for 12 h. Chromatin immunoprecipitation was performed with anti-Myc antibody prior to PCR amplification. Cellular DNA was used as the input control. D, schematic model for PIAS1-directed repression of LXR-dependent gene transcription activation. PIAS1 can interact with LXRs as well as PGC-1 co-activators to disrupt their association. Without relying on the SUMO-modifications of LXRs, PIAS1 is able to block LXR binding to the promoters of their target genes (e.g. Srebp1c). Thus, PIAS1 acts to dampen ligand-activated, LXR-dependent gene expression programs such as lipogenesis.
