Background: Pentatricopeptide repeat (PPR) proteins act at specific RNA-editing sites in plant mitochondria.
Results: Two related PPR proteins with only five repeat units differentially influence RNA editing in pollen and leaves.
Conclusion: Two PPR proteins target the same two sites in RNA editing.
Significance: This is the first report on overlapping specificities of PPR proteins in RNA editing.
Keywords: Mitochondria, Plant Molecular Biology, RNA-binding Protein, RNA Editing, RNA Processing, PPR Protein
Abstract
The facilitators for specific cytosine-to-uridine RNA-editing events in plant mitochondria and plastids are pentatricopeptide repeat (PPR)-containing proteins with specific additional C-terminal domains. Here we report the related PPR proteins mitochondrial editing factor 8 (MEF8) and MEF8S with only five such repeats each to be both involved in RNA editing at the same two sites in mitochondria of Arabidopsis thaliana. Mutants of MEF8 show diminished editing in leaves but not in pollen, whereas mutants of the related protein MEF8S show reduced RNA editing in pollen but not in leaves. Overexpressed MEF8 or MEF8S both increase editing at the two target sites in a mef8 mutant. Double mutants of MEF8 and MEF8S are not viable although both identified target sites are in mRNAs for nonessential proteins. This suggests that MEF8 and MEF8S may have other essential functions beyond these two editing sites in complex I mRNAs.
Introduction
RNA editing in mitochondria of flowering plants changes 400–500 selected cytosines to uridines mostly in coding regions of mRNAs and some tRNAs (1–3). Specific sequence contexts in the pre-mRNA act as cis-elements that distinguish an editing site from a cytosine remaining unedited (4–7). In Arabidopsis thaliana, these RNA sequences are presumably recognized by nuclear-encoded specificity factors such as mitochondrial editing factor 1 (MEF1),2 MEF9, MEF11, MEF14, and MEF18–MEF22, which are required for correct editing of specific sites (8–13).
The generally one-to-one relationship between specific cis-elements in the mitochondrial RNA molecules and individual trans-factors, the MEF proteins, is most parsimoniously explained by the MEF proteins acting directly as RNA-binding proteins. This speculation is supported by the finding that some MEF proteins are required for editing at several sites that are preceded by similar cis-elements. Direct investigations of an analogous plastid RNA-editing factor, the CRR4 protein, have shown that this protein indeed binds to the specific RNA sequences at its cognate RNA-editing site (14). Related proteins involved in RNA processing such as PPR5 and PPR10 (15, 16) and others of unknown function (17) also bind to specific RNA sequences or at least contact polyribonucleotides.
The proteins required for specific RNA-editing events in plastids and in mitochondria, including for example CRR4, the MEFs, REME1 (18), and OGR1 in rice (19), are all pentatricopeptide repeat proteins (PPR proteins). Those identified to date are characterized by containing three types of repeats that vary between 31 amino acids in short, 35 in medium, and 37 in long repeat elements and contain an additional extension (E) domain. The single exception so far is a PPR protein with only medium-type repeats and no C-terminal extension; the absence of this protein enhances editing at several sites (20). A number of the mitochondria- and plastid-targeted RNA-editing PPR proteins are extended beyond the E domain by an additional approximately 100 amino acids long DYW region with the name-giving amino acid triplet DYW at the C terminus whereas others end with the E domain (18, 21–29). Direct investigations have shown that in some editing proteins, the DYW domain is essential but can be deleted in others (23, 30, 31). The presence of conserved features of cytidine deaminases in most DYW domains prompted speculations that these may contribute the as-yet-unidentified enzymatic activity (32), but an experimental investigation so far found only an RNA-degrading activity (33).
If the RNA-editing factors in the E subclass of the approximately 450 PPR proteins encoded in the nuclear genome of Arabidopsis (17, 34–37) indeed interact directly with specific RNA motifs at their cognate RNA-editing sites, one would expect that the loss of a given RNA-editing PPR protein would lead to the loss of editing at these target sites. This is in fact observed for many of these proteins, suggesting that there are usually no back-up factors that can substitute these specific functions (e.g. 8–13, 24, 25). In instances of partial residual editing in an apparent knockout of a given MEF protein (9, 12), another at least partially substituting editing factor must be postulated, but none has been identified.
We here report the identification of two novel PPR proteins of the DYW class with fewer PPRs than previously identified factors. These related MEF8 and MEF8S proteins are involved in RNA editing at the same two specific sites in plant mitochondria.
EXPERIMENTAL PROCEDURES
Plant Material and Preparation of Nucleic Acids
A. thaliana seeds of wild type Columbia (Col) and the various mutants were grown as described (38). DNA or RNA from the leaves of the A. thaliana plants were prepared by published procedures (39). For pollen analysis, pollen was collected from flower buds just prior to their opening by manually opening them inside an Eppendorf tube. Pollen shaken out attached electrostatically to the wall of the tube. Usually pollen from seven flowers was collected. For RNA preparations, lysis buffer from commercial RNA kits (GE Healthcare) was added, and pollen was frozen and thawed twice.
Identification of the mef8 Mutants
Mutant mef8-1 was identified in a screen of a population of ethyl methanesulfonate (EMS) mutagenized plants by its reduced editing at site nad5-676 (40–42). In parallel to the mapping of the gene mutated in this EMS plant, a screen of T-DNA insertion mutants for altered RNA editing identified site nad5-676 as a target of the locus At2g25580 (9). This mutant was accordingly named mef8-2.
Analysis of RNA-editing Sites
Specific cDNA fragments were generated by RT-PCR amplification following established protocols (39). The cDNA sequences were compared for C-to-T differences resulting from RNA editing. Most sequences were obtained commercially from 4base lab (Reutlingen, Germany), LGC Genomics (Berlin, Germany), or from Macrogen (Seoul, Korea). Evaluation of sequence data was done by measuring peak heights. Percentages of editing were obtained as the relationship of the peak height of the T signal to the sum of the T and C signals at the respective editing site (12).
Stable Transformation of Plants
Plants of the T-DNA mutant line mef8-2 were transformed by floral dip (43) with the MEF8 or MEF8S wild type Col reading frame under control of the 35S promoter in vector pMDC123 (44). Transgenic plants were selected by spraying with Basta®. For the analysis of RNA-editing levels, the respective cDNA fragments were sequenced, and relative peak heights were compared (12, 38).
RESULTS
Identification of MEF8 as a Factor of RNA Editing at Site nad5-676
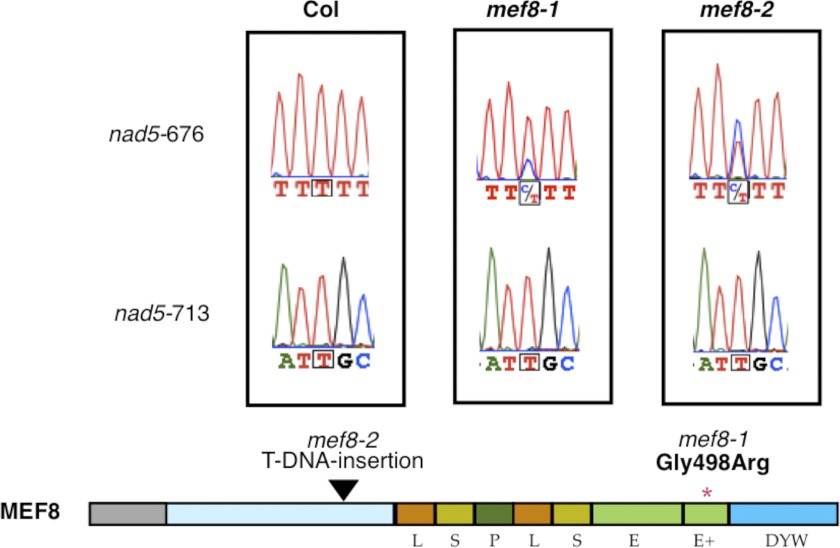
With the recently developed multiplexed SNaPshot approach, 369 annotated editing sites were probed in 2,000 individuals of a population of EMS mutant plants to directly find mutants impaired in RNA editing at one or more of the investigated sites in plant mitochondria (40–42). This screen for deficiencies in RNA editing at specific sites identified a plant with reduced editing at site nad5-676. The homozygous mutant plant shows editing at this site to be diminished to approximately 70% in comparison with the wild type Columbia plants in which this site is altered to 100% from the genomically encoded C to an U (Fig. 1). The genomic locus responsible for this reduction was mapped by crosses between the mutant plants and wild type plants of ecotype Ler to the site annotated At2g25580. This gene, now named MEF8, encodes the editing specificity factor MEF8.
FIGURE 1.
Two independent mutants of the gene for MEF8 show reduced RNA-editing levels at the same site in the mitochondrial nad5 mRNA. The mef8-1 EMS-induced A. thaliana mutant plant was identified in a cDNA SNaPshot analysis by its lower C-to-U-editing level at the mitochondrial nad5-676 RNA-editing site. Direct sequence analysis shows that the editing level is reduced from 100% in Col wild type plants to approximately 70%. In a second mutant plant with a T-DNA insertion, mef8-2 (Salk_106391), a stronger reduction to approximately 40% is seen. In contrast, another site in the same mRNA, site nad5-713, is correctly edited to completion in wild type and in mutant plants. Color traces are C, blue; T, red; G, black; A, green. The lower part shows the predicted structure of the MEF8 protein. The different types of repeats, the E, E+, and DYW elements, the predicted mitochondrial import sequence, and the unstructured region (light blue) are color-coded. The location of the EMS point mutation and the resulting change of a glycine-to-an arginine codon in mef8-1 and the location of the T-DNA insertion in mef8-2 are indicated.
To corroborate the connection between the EMS mutant line (now named mef8-1) and the editing defect, we next analyzed an independent mutation in MEF8, T-DNA insertion line SALK_106391 (now named mef8-2), for editing at the target site (Fig. 1). In this plant, editing at site nad5-676 is reduced to approximately 40%, confirming the involvement of MEF8 in RNA editing at this site. As a control the closest RNA-editing site in the same mRNA, site nad5-713 was analyzed (Fig. 1). This site is edited to apparent completion in both mutants as well as in WT plants, excluding any secondary influence on RNA editing by altered turnover or transcription rates and suggests that MEF8 is specifically involved in editing at site nad5-676.
The difference in residual editing in the two mutants (70 and 40%, respectively) suggests that the function of the mutant mef8-1 protein is only partially inhibited. In mutant mef8-2 presumably no functional MEF8 protein is made, which reduces the remaining editing activity more strongly. If this scenario is correct, another factor must be responsible for the 40% remaining editing in mutant mef8-2.
A Second RNA-editing Site Is Targeted by MEF8
Sequence comparison of the target site nad5-676 in several plant species shows that this editing event of A. thaliana is conserved in only few species such as Brassica napus, whereas most other flowering plants code for a U at this position already in the genome. The amino acid phenylalanine at this position and the surrounding amino acids in the NAD5 protein are highly conserved, the only exception being Oenothera berteriana, where editing at this C has not been observed and a leucine may be encoded instead. Alternatively, the editing event may have been missed in this plant.
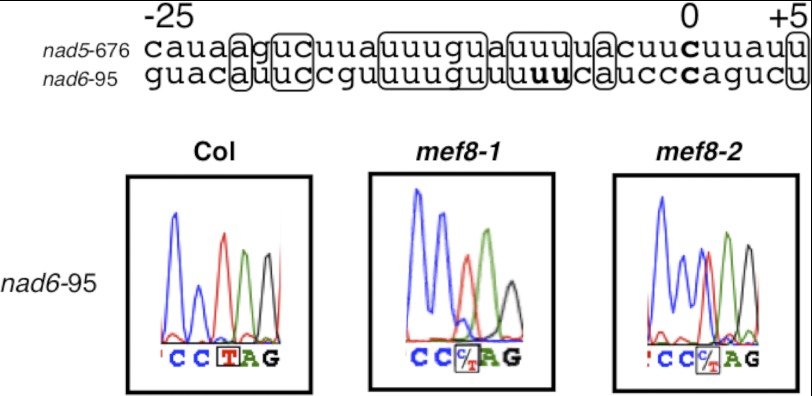
An in silico search of the mitochondrial transcriptome sequence of A. thaliana with the presumed cis-element of the nad5-676 target site yields the upstream region of editing site nad6-95 as the most similar sequence pattern (Fig. 2, upper part). Investigation of RNA editing at this site indeed shows an effect in leaves of the mef8 mutants, editing being reduced from 100% in wild type RNA to approximately 80% in mutant mef8-1 and to about 50% in mutant mef8-2 (Fig. 2, bottom part).
FIGURE 2.
A second site with a similar cis-sequence is also affected in the mef8 mutants. The upper part shows an alignment of the nad5-676 target site with the nad6-95 site with a similar cis-element in the region −25 to +5 relative to the edited C nucleotide (bold C at nucleotide 0). Nucleotides derived from other editing events are given as bold U, nucleotides identical between the two sequences are framed. The target editing site is labeled 0. The lower part shows a cDNA sequence analysis of the second site nad6-95, at which editing is reduced to approximately 80% in the mef8-1 and to 50% in the mef8-2 mutant from the nearly 100% editing observed in wild type plants.
A Gene Similar to the MEF8 Gene in the Arabidopsis Genome
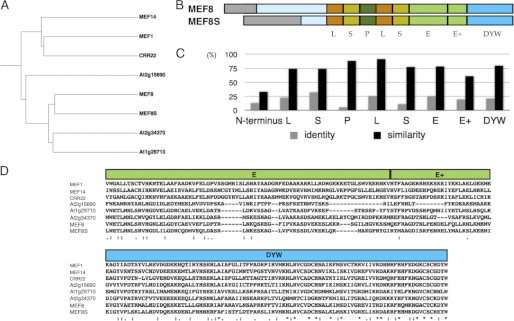
The conclusion outlined above that another factor has to be able to compensate at least partially for the loss of MEF8 extends to the two editing sites, because both target sites show residual editing even in the T-DNA insertion line mef8-2. In this line the MEF8 protein is presumably disrupted and another factor must provide the residual activity. One possible candidate for such an alternative factor might be a protein with characteristics similar to MEF8. An in silico search in A. thaliana for genes encoding proteins similar to MEF8 identified a genomic locus (At4g32450) that codes for a protein in which overall approximately 25% of the amino acids are identical, this conservation increasing up to 80% similarity in some repeats (Fig. 3). Three related proteins with structures similar to the MEF8/MEF8S pair are encoded in the Arabidopsis genome, the two proteins encoded at loci At2g34370 and At1g29710 form another pair, and the one encoded at locus At2g15690 is more distantly related (Fig. 3A).
FIGURE 3.
Two similar proteins, MEF8 and MEF8S, are encoded in the A. thaliana nuclear genome. A, a similarity tree with the MEF8 and MEF8S protein pair, three other proteins with related features, and the mitochondrial MEF14, MEF1, and the plastid CRR22 PPR proteins. This comparison shows that the two proteins encoded by At2g34370 and At1g29710 are closely related to each other and also similar to the MEF8/MEF8S pair. The PPR protein encoded by At2g15690 likewise has few PPRs and higher primary sequence similarity to these than to other PPR proteins encoded in the Arabidopsis genome. Sequences were aligned with the Clustal W program in the UniProt database. B, alignment of the schematic structure of the MEF8 and MEF8S PPR proteins encoded by locus At2g25580 and At4g32450, respectively. The N-terminal amino acid sequences are predicted to contain mitochondrial target peptides (gray) followed by amino acid stretches with no clear structure (light blue) before the first detectable PPRs. According to their sizes and conserved features, five PPR elements can be discerned as indicated. Color coding is as described in the legend to Fig. 1. C, percentages of amino acid identity (gray bars) and similarity (black bars) between the MEF8 and MEF8S proteins are given for each of the repeats, the N-terminal region up to the first repeat and the E, E+, and DYW elements. The high degree of overall similarity suggests a common evolutionary origin of both genes and also a functional equivalence. D, amino acid alignment of the E, E+, and DYW elements of the MEF8 and MEF8S proteins with the respective sequences from the three similar proteins and from the editing factors MEF1 (12), MEF14 (11), and CRR22 (55) compared in A shows the gaps in the E domain. The C-terminal DYW triplet is altered to EYW in MEF8S.
The MEF8 protein (and these related proteins) is unique in comparison with the other so far identified PPR proteins involved in RNA editing in that it contains an extremely reduced repeat domain with only five repeats (Figs. 1 and 3). Some of these repeats lack recognizable conservation of the amino acid moieties characteristic for PPRs and could therefore only be delineated by alignment with consensus patterns including evolutionary far distant species such as Naegleria (45). Degeneration is also seen in the E−/E+ region, including a >10 residues deletion. The MEF8-like protein ends with the EYW triplet instead of the highly conserved DYW motif.
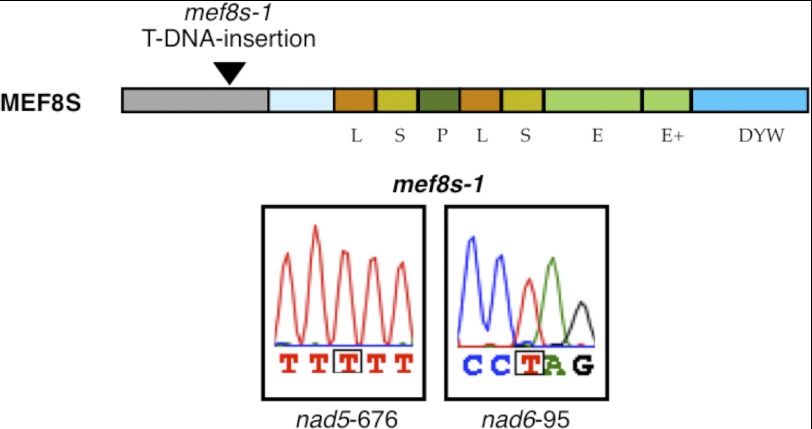
For its high similarity, we renamed this MEF8-like protein MEF8S. To investigate the potentially similar function of the MEF8S protein, we analyzed RNA editing at the MEF8 target sites in a T-DNA insertion mutant of the MEF8S gene locus, now named mef8s-1 (SALK_047005C; Fig. 4). Disruption of MEF8S has no detectable effect on processing of these sites in leaves, because, like in wild type plants, both sites are edited apparently completely in the steady-state mRNA population. Furthermore, 369 documented RNA-editing sites analyzed in leaves of the mef8s-1 mutant by the SNaPshot procedure are all edited as in wild type plants.
FIGURE 4.
Editing at the MEF8 target sites is not affected in leaves of a MEF8S mutant. The top part shows the mef8s-1 mutant with a T-DNA insertion in the open reading frame coding for the N-terminal region predicted as mitochondrial target sequence of the MEF8S protein. The sequence traces below show that both MEF8 target sequences are fully edited in leaves of the homozygous mef8s-1 mutant plants.
Complementation of the MEF8 T-DNA Insertion Mutant
The connection between the MEF8 gene and the reduced RNA editing at the nad5-676 and nad6-95 target sites was further assayed by exploring the ability of the WT Col MEF8 and MEF8S genes to complement the mef8-2 T-DNA insertion mutant. Transfection of the MEF8 gene into mef8-2 mutant protoplasts increased RNA editing significantly (data not shown). Protoplast complementation with the MEF8S gene also shows a slight increase of RNA editing at both sites that is however statistically not significant (data not shown).
To investigate this observation further, we stably transformed the mef8-2 T-DNA insertion mutant with either the MEF8 or the MEF8S gene under control of the 35S promoter to achieve (relatively) high levels of expression (Fig. 5). In the transgenic plants, the low levels of RNA editing at the target sites, nad5-676 and nad6-95, are significantly increased in leaves by either the MEF8 or the MEF8S gene. In both instances, recovery is better by MEF8 than by MEF8S.
FIGURE 5.

Complementation of the T-DNA mutant line mef8-2 by stable transformation with the Col MEF8 or the Col MEF8S gene. Sequence tracings of the cDNA analysis from stable transformants of the T-DNA mutant line mef8-2 with the Col MEF8 gene or the Col MEF8S gene under control of the 35S promoter show that both genes increase RNA-editing levels in leaves. The effect of MEF8S is greater at the nad6-95 editing site than at site nad5-676. The bars show data from two independently derived transgenic plants, the S.D. is indicated.
Double Knockout of MEF8 and MEF8S Is Embryo-lethal
To further investigate the complementing functions of MEF8 and MEF8S in editing the two MEF8 target sites, we tried to establish a double knockout of MEF8 and MEF8S. We crossed the two homozygous T-DNA insertion lines of MEF8 and MEF8S and analyzed the offspring for the presence of the mutant genes. Surprisingly, we did not obtain any viable offspring in repeated crossings. No seeds developed, and no embryo grew detectably.
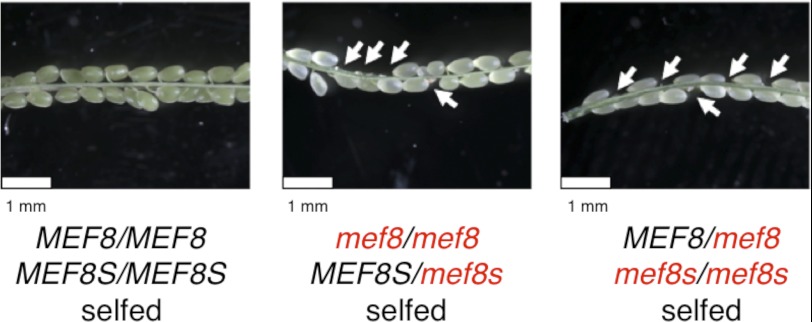
This is unexpected because the single mutants mef8-2 and mef8s-1 do not show any gross abnormalities in their growth habits. To exclude any problems potentially particular to the combination of the two homozygous mutant plants, we investigated the offspring of two different crosses (Fig. 6): For the first, plants homozygous for mef8-2 and heterozygous for mef8s-1 were generated by crossing the homozygous mutant of mef8-2 with a plant heterozygous at mef8s-1 and selection of respective individuals by PCR for the presence of the two T-DNA insertions. The second plant line was analogously obtained and is homozygous for mef8s-1 and heterozygous for mef8-2. Both plant lines were selfed, and 160 randomly chosen individuals, respectively, were screened for offspring homozygous for both mutations. None was identified.
FIGURE 6.
The combination of homozygous disabled mef8 and mef8s alleles is embryo-lethal. Self-pollinations of plants with either homozygous mef8-2 and heterozygous mef8s-1 alleles (center panel) or of plants with homozygous mef8s-1 and heterozygous mef8-2 loci (right panel) show approximately 25% aborted seeds in the opened siliques (arrows). None of the approximately 160 plants recovered and analyzed of each cross is homozygous for both mutant genes. In wild type Col selfings, all seeds develop normally (left panel). Disabled alleles of the genes indicated are marked in red, intact alleles in black in the schematic below the photographs.
To see if (and if so at what stage) embryo development was compromised, seed pods of both crosses were analyzed (Fig. 6). Whereas control wild type selfings showed nearly full pods, each of the two crosses revealed approximately 25% aborted seed sites. The phenotypic appearance is very similar to embryo-lethal mutants as classified and described in detail (46, 47).
Counterregulated Expression of the MEF8 and MEF8S Genes in Different Tissues Results in Distinct RNA-editing Phenotypes in Their Mutants
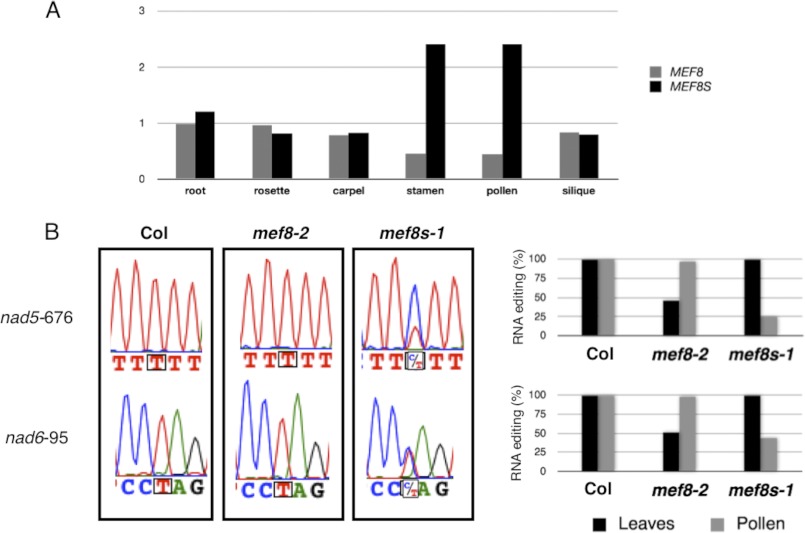
The Atgenexpress analysis of MEF8 and MEF8S gene expression patterns shows the overall very low level of transcription typical for MEF genes throughout the various tissues and growth conditions 48). The most striking differences between the steady-state transcript levels in distinct tissues between these two similar genes are seen in the floral organs (Fig. 7A). Whereas the transcript levels of MEF8S are elevated approximately 3-fold in stamen and pollen, the amount of steady-state transcripts of MEF8 is reduced to about half of the level in other tissues. This observation suggests a potentially stronger influence of MEF8S during pollen development than that of MEF8.
FIGURE 7.
RNA-editing levels at the MEF8 and MEF8S target sites in pollen and in leaves of the respective knock-out mutants correspond with the expression levels of the remaining MEF8S and MEF8 genes. A, Atgenexpress data (48) show that MEF8S expression levels (measured as quantities of steady-state RNA signals) increase in stamen tissues and in pollen, whereas transcript levels of MEF8 are lower in these cells than in any other plant tissue. B, pollen kernels were collected from still closed flower buds, and the cDNA obtained from the young pollen was analyzed for editing at the two target sites. Pollen from the homozygous mutant plant mef8-2 with intact MEF8S alleles (center sequencing panels; gray bars in histograms on the right) is hardly affected, and editing levels are similar to those in pollen from wild type Col plants (left sequencing panel). In leaves, editing at both target sites is reduced (black bars in histograms on the right; Figs. 1 and 2). Editing in pollen from the homozygous mutant plant mef8s-1 (right sequencing panels; gray bars in histograms on the right) is severely reduced whereas editing in leaves is unaffected. In pollen, the intact MEF8 alleles cannot compensate the loss of MEF8S, possibly because of the low expression of MEF8 in these tissues.
To investigate this possibility, we compared the RNA-editing levels at the two target sites in pollen obtained from nearly mature but still closed flower buds from the three different genotypes: wild type plants, plants homozygous for mef8-2, and plants homozygous for mef8s-1 (Fig. 7B). The average level of editing in pollen from plants without a functional MEF8 gene, i.e. those homozygous for mef8-2, was similar to that in wild type plants and hardly affected. On the other hand, pollen from plants homozygous for mef8s-1 without a functional MEF8S gene showed reduced levels of editing at both target sites (Fig. 7B).
The comparison of editing in leaves and in pollen shows that plants homozygous for mef8-2 show the phenotype of reduced editing only in leaves but not in pollen, whereas plants homozygous for mef8s-1 have reduced RNA editing in pollen but not in leaves (Fig. 7B).
Pollen kernels were analyzed for viability by the Alexander stain. All kernels from both mutants showed a percentage of positive staining identical to the wild type, suggesting that the altered mitochondrial RNA editing does not manifest in the phenotypic appearance and in the viability of the pollen as such.
DISCUSSION
RNA-editing Proteins with a Degenerated PPR Protein Skeleton
The two PPR proteins MEF8 and MEF8S are unusual in comparison with the previously assigned editing factors of mitochondria and plastids: both contain very short PPR domains with only five PPRs (Fig. 3). The assigned five medium, long, and short elements in MEF8 and MEF8S furthermore deviate from the characteristic amino acid signature with several of the usually conserved residues substituted by unconventional moieties (17, 45, 49, 50). The E domain shows a deletion of 11 amino acids in MEF8 and of 13 residues in MEF8S in comparison with the E domain consensus arrangement (Fig. 3D). The adjacent E+ region and the following DYW domain are somewhat better maintained and have retained most conserved key elements.
Three further proteins with related features of few PPRs, a shorter E domain and high primary sequence similarity are present in the Arabidopsis genome (Fig. 3). Similar to the MEF8 and MEF8S pair, the two proteins encoded by At2g34370 and At1g29710 form another pair more closely related to each other than to other PPR proteins. The presence of shorter E and DYW domains in the three MEF8/MEF8S related proteins suggests that they may also be involved in RNA editing, but this will have to be investigated in detail.
MEF8 and MEF8S Proteins with Only Five PPRs Are Site-specific
The restricted number of PPRs in the MEF8 and MEF8S proteins raises the question of how this fits with the current model idea that the PPRs recognize and bind to a specific RNA sequence on a one-on-one basis, i.e. one repeat element attaching to one nucleotide. Precedence for such a connection between α-helical 35-amino acid units and individual nucleotides is found in several DNA-binding proteins, such as the TAL regulators (51).
With only a maximum of five such repeat units in the MEF8 and MEF8S proteins, even binding of all five repeats to specific nucleotides would statistically not be sufficient to yield unique interactions within the transcriptome of the 367-kb large mitochondrial genome of Arabidopsis (52). To achieve maximal specificity, all of the repeats in the MEF8 and MEF8S proteins should contact individual nucleotides in the target RNA. In larger RNA-editing specificity factors, the repeats may not all bind to nucleotides, but some may fulfill a function as spacer in the PPR proteins to allow gaps in the contacted nucleotide sequences. When several RNA target sites are addressed by individual PPRs, these RNAs often reveal some sort of consensus pattern only if gaps are allowed (12, 23, 30, 53–56).
Other proteins with similarly rather short tracts have been found to interact with RNA. The plant mitochondrial RNase P (57) recognizes tRNA structures and processes these at specific sites. The THA8 protein with only four copies of PPRs nevertheless binds to specific introns in the plastid and is essential for splicing of these sequences (58).
Both target sites of MEF8 and MEF8S are rather U-rich (Fig. 2), and at least some of the repeats contacting the RNA should be specific for U-nucleotides. A detailed analysis of the interaction of these unique five PPRs in MEF8 and MEF8S proteins with their RNA targets may provide experimental access to the mode and parameters of protein-RNA recognition by the RNA-editing PPR proteins.
MEF8 and MEF8S Proteins Target the Same Editing Sites
The initial identification of the EMS mutant mef8-1 with lowered but not abolished editing can be interpreted as partial inactivation of the MEF8 protein by the single amino acid exchange in the E+ domain (Fig. 1). The stronger reduced mitochondrial editing in the T-DNA insertion mutant mef8-2, which interrupts the MEF8 protein within the N-terminal region (Fig. 1), suggests that the single amino acid exchange in mef8-1 does indeed disturb MEF8 function but does not abolish it completely. The T-DNA insertion in the MEF8 reading frame in mef8-2, on the other hand, should prohibit expression of a functional MEF8 protein. Therefore, the observed residual editing in this mutant requires another factor to compensate for the destroyed MEF8 protein and to fulfill its role at least partially. The similar PPR protein MEF8S is a candidate for this second factor.
One crucial condition is that MEF8S targets the same editing sites as MEF8. This is confirmed by several lines of evidence. Stable transformation of mef8-2 mutant plants with either MEF8 or MEF8S increases the rate of editing at both target sites significantly (Fig. 5). The positive effect of the transformation with MEF8S indicates that this protein is indeed involved in editing of these sites. This result furthermore suggests that the low level of expression of MEF8S in leaves limits compensation of the loss of MEF8 in untransformed mef8-2 mutant plants (Fig. 7).
The tissue-specific RNA-editing phenotype in the MEF8S knock-out mutant plant further confirms that MEF8 and MEF8S target the same editing sites. RNA editing at the MEF8 target sites is reduced in pollen from the mef8s-1 mutant (Fig. 7B). The MEF8 level of expression in leaves in this mutant is sufficient to accommodate the loss of MEF8S, but the reduced level of MEF8 expression in pollen is not enough to compensate for the absence of MEF8S. These coinciding patterns of expression and RNA-editing levels (Fig. 7) confirm that both proteins are involved in editing the same two sites and that the expression levels of the mef8 and mef8s genes limit the efficiency of RNA editing at these two sites.
These results confirm that the two proteins MEF8 and MEF8S can substitute for each other at least partially in RNA editing. Either MEF8 or MEF8S can presumably bind to the two target sequences and connect to other proteins in the hypothetical editosome, possibly a multiple organellar RNA-editing factor protein (59). Unfortunately, the substitution effect cannot be tested directly in a double mutant of mef8 and mef8s because we find that this is not viable.
MEF8 and MEF8S Must Have Additional, Essential Functions
The observation that homozygous plants mutated in both MEF8 and MEF8S genes are not viable remains as yet unexplained (Fig. 6). The embryo-lethal double knockout suggests that the two proteins can substitute for each other at one or more functions that are essential for survival of the plant. It is unlikely that the two MEF8 and MEF8S RNA-editing target sites in mRNAs for complex I subunits, nad5-676 and nad6-95, are these crucial functions because plants without functional complex I are viable.
Therefore, additional RNA-editing sites that are essential for survival of the embryo are likely to be targeted. These have not yet been identified, and further investigations will be required to answer this question. Mutations in some PPR proteins and the concomitant loss of their respective RNA-editing events in plastids or mitochondria result in severe phenotypes. The loss of a large number of editing sites has been found to be likewise embryo-lethal as homozygous T-DNA mutants of the mitochondrial editing cofactor multiple organellar RNA-editing factor 1 are not viable (59). Both crosses from self-pollinated plants, those homozygous for mef8-2 and heterozygous for mef8s-1 and those homozygous for mef8s-1 and heterozygous for mef8-2, exhibit an embryo-defective phenotype in approximately 25% of the seed pods in immature siliques (Fig. 6). Similar embryo-lethal phenotypes have been observed with knock-out mutants of mitochondrial or chloroplast aminoacyl-tRNA-synthetases (46, 47). Accordingly, MEF8 and MEF8S seem to be required for proper development of the embryo.
Alternatively, it is possible that MEF8 and MEF8S have additional other essential functions beyond their involvement in RNA editing. Yet another explanation may be that either the NAD5 or the NAD6 proteins derived from mRNAs not edited at the nad5-676 and nad6-95 target sites of MEF8 and MEF8S result in mutant proteins that interfere with essential mitochondrial functions. It will be interesting to clarify the potential additional function of MEF8 and MEF8S and to determine whether indeed the specific RNA-binding preferences of the two proteins have begun to drift apart since their presumed separation.
Are the Genes for MEF8 and MEF8S Derived by Duplication?
Current models for the evolution of the PPR proteins involve an ancestral gene of the medium-long-short class to which E and DYW domains had been added (32, 34, 36). These original medium-long-short-E-DYW arrangements are the only editing factors in the moss Physcomitrella patens (49, 50, 53, 60). Subsequently, this ancestral gene was amplified, and the resulting large family of genes allowed establishment of the many RNA-editing sites observed in flowering plants and even more so in Lycopodium (61).
The MEF8 and MEF8S protein-coding genes most likely arose by such a gene duplication. This duplication has presumably taken place rather recently in evolution because both proteins are still recognizably similar and still target the same RNA sequence although possibly with slightly differing preferences. Furthermore, in other plant genomes, two genes coding for proteins similar to MEF8 and MEF8S are detected only in Arabidopsis lyrata, whereas in more distant plant species only single similar genes are retrieved.
These potential orthologs in vine, poplar, and rice match the consensus structure of MEF8 and MEF8S, with only five PPRs in the central part. These proteins show a similar degeneration of the C-terminal part of the E domain with an analogous deletion after the conserved N-terminal region.
Prediction programs suggest 64 amino acids for MEF8 and 110 residues for MEF8S as organellar target sequences with predicted mitochondrial locations. These rather long presequences leave unique N-terminal regions in both proteins up to the first PPRs for which little structural features are discerned. Their potential function in binding to RNA target sequences and in recruiting other editing factors needs to be determined experimentally.
In summary, our findings show that the two similar RNA-editing factors MEF8 and MEF8S can (partially) substitute for each other. This proves previous indirect surmises about additional editing factors acting in instances of partial editing mutants. In presumed knock-out mutants of MEF1, for example, two sites have completely lost editing whereas the third target is still edited to 20% (12). Substitution of the activity by another factor has been proposed, but none had been identified previously. The two PPR proteins MEF8 and MEF8S with only few PPRs and their presumed continuous contact to their RNA target sequences may allow direct advances toward deciphering the PPR-RNA code in plant organelles.
Acknowledgments
We thank Mareike Rüdinger and Volker Knoop at Molekulare Evolution, Universität Bonn, for analysis of the repeat structure and number in MEF8 and MEF8S; Rita Gross-Hardt at the ZMBP, Universität Tübingen, for help with the interpretation of the embryo-lethal phenotypes; Stefan Britsch for access to microscope facilities and Dagmar Pruchner and Angelika Müller for excellent experimental help.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (to M. T. and A. B.).
- MEF
- mitochondrial editing factor
- PPR
- pentatricopeptide repeat
- Col
- Columbia
- EMS
- ethyl methanesulfonate.
REFERENCES
- 1. Giegé P., Brennicke A. (1999) RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. U.S.A. 96, 15324–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Handa H. (2003) The complete nucleotide sequence and RNA-editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 31, 5907–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takenaka M., Verbitskiy D., van der Merwe J. A., Zehrmann A., Brennicke A. (2008) The process of RNA editing in plant mitochondria. Mitochondrion 8, 35–46 [DOI] [PubMed] [Google Scholar]
- 4. Bock R., Hermann M., Kössel H. (1996) In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J. 15, 5052–5059 [PMC free article] [PubMed] [Google Scholar]
- 5. Farré J. C., Leon G., Jordana X., Araya A. (2001) cis-Recognition elements in plant mitochondrion RNA editing. Mol. Cell. Biol. 21, 6731–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neuwirt J., Takenaka M., van der Merwe J. A., Brennicke A. (2005) An in vitro RNA-editing system from cauliflower mitochondria: editing site recognition parameters can vary in different plant species. RNA 11, 1563–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van der Merwe J. A., Takenaka M., Neuwirt J., Verbitskiy D., Brennicke A. (2006) RNA-editing sites in plant mitochondria can share cis-elements. FEBS Lett. 580, 268–272 [DOI] [PubMed] [Google Scholar]
- 8. Takenaka M. (2010) MEF9, an E-subclass pentatricopeptide repeat protein, is required for an RNA-editing event in the nad7 transcript in mitochondria of Arabidopsis. Plant Physiol. 152, 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takenaka M., Verbitskiy D., Zehrmann A., Brennicke A. (2010) Reverse genetic screening identifies five E-class PPR proteins involved in RNA editing in mitochondria of Arabidopsis thaliana. J. Biol. Chem. 285, 27122–27129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Verbitskiy D., Zehrmann A., van der Merwe J. A., Brennicke A., Takenaka M. (2010) The PPR protein encoded by the lovastatin insensitive 1 gene is involved in RNA editing at three sites in mitochondria of Arabidopsis thaliana. Plant J. 61, 446–455 [DOI] [PubMed] [Google Scholar]
- 11. Verbitskiy D., Härtel B., Zehrmann A., Brennicke A., Takenaka M. (2011) The DYW-E-PPR protein MEF14 is required for RNA editing at site matR-1895 in mitochondria of Arabidopsis thaliana. FEBS Lett. 585, 700–704 [DOI] [PubMed] [Google Scholar]
- 12. Zehrmann A., Verbitskiy D., van der Merwe J. A., Brennicke A., Takenaka M. (2009) A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell 21, 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zehrmann A., Verbitskiy D., Härtel B., Brennicke A., Takenaka M. (2011) PPR proteins network as site-specific RNA-editing factors in plant organelles. RNA Biol. 8, 67–70 [DOI] [PubMed] [Google Scholar]
- 14. Okuda K., Nakamura T., Sugita M., Shimizu T., Shikanai T. (2006) A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J. Biol. Chem. 281, 37661–37667 [DOI] [PubMed] [Google Scholar]
- 15. Beick S., Schmitz-Linneweber C., Williams-Carrier R., Jensen B., Barkan A. (2008) The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 28, 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pfalz J., Bayraktar O. A., Prikryl J., Barkan A. (2009) Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28, 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lurin C., Andrés C., Aubourg S., Bellaoui M., Bitton F., Bruyère C., Caboche M., Debast C., Gualberto J., Hoffmann B., Lecharny A., Le Ret M., Martin-Magniette M. L., Mireau H., Peeters N., Renou J. P., Szurek B., Taconnat L., Small I. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bentolila S., Knight W., Hanson M. (2010) Natural variation in Arabidopsis leads to the identification of REME1, a pentatricopeptide repeat-DYW protein controlling the editing of mitochondrial transcripts. Plant Physiol. 154, 1966–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim S. R., Yang J. I., Moon S., Ryu C. H., An K., Kim K. M., Yim J., An G. (2009) Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. Plant J. 59, 738–749 [DOI] [PubMed] [Google Scholar]
- 20. Doniwa Y., Ueda M., Ueta M., Wada A., Kadowaki K., Tsutsumi N. (2010) The involvement of a PPR protein of the P subfamily in partial RNA editing of an Arabidopsis mitochondrial transcript. Gene 454, 39–46 [DOI] [PubMed] [Google Scholar]
- 21. Chateigner-Boutin A. L., Ramos-Vega M., Guevara-García A., Andrés C., de la Luz, Gutiérrez-Nava M., Cantero A., Delannoy E., Jiménez L. F., Lurin C., Small I., León P. (2008) CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56, 590–602 [DOI] [PubMed] [Google Scholar]
- 22. Fujii S., Small I. (2011) The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 191, 37–47 [DOI] [PubMed] [Google Scholar]
- 23. Hammani K., Okuda K., Tanz S. K., Chateigner-Boutin A. L., Shikanai T., Small I. (2009) A study of new Arabidopsis chloroplast RNA-editing mutants reveals general features of editing factors and their target sites. Plant Cell 21, 3686–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kotera E., Tasaka M., Shikanai T. (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330 [DOI] [PubMed] [Google Scholar]
- 25. Okuda K., Myouga F., Motohashi R., Shinozaki K., Shikanai T. (2007) Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. U.S.A. 104, 8178–8183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robbins J. C., Heller W. P., Hanson M. R. (2009) A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA 15, 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shikanai T. (2006) RNA editing in plant organelles: machinery, physiological function and evolution. Cell Mol. Life Sci. 63, 689–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu Q. B., Jiang Y., Chong K., Yang Z. N. (2009) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J. 59, 1011–1023 [DOI] [PubMed] [Google Scholar]
- 29. Zhou W., Cheng Y., Yap A., Chateigner-Boutin A. L., Delannoy E., Hammani K., Small I., Huang J. (2009) The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 58, 82–96 [DOI] [PubMed] [Google Scholar]
- 30. Verbitskiy D., Zehrmann A., Brennicke A., Takenaka M. (2010) The DYW domain in the MEF11 RNA-editing protein seems to be involved in specific RNA binding rather than in the enzymatic reaction. Plant Signal. Behav. 5, 558–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zehrmann A., Verbitskiy D., Härtel B., Brennicke A., Takenaka M. (2010) RNA editing competence of trans-factor MEF1 is modulated by ecotype-specific differences but requires the DYW domain. FEBS Lett. 584, 4181–4186 [DOI] [PubMed] [Google Scholar]
- 32. Salone V., Rüdinger M., Polsakiewicz M., Hoffmann B., Groth-Malonek M., Szurek B., Small I., Knoop V., Lurin C. (2007) A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 581, 4132–4138 [DOI] [PubMed] [Google Scholar]
- 33. Nakamura T., Sugita M. (2008) A conserved DYW domain of the pentatricopeptide repeat protein possesses a novel endoribonuclease activity. FEBS Lett. 582, 4163–4168 [DOI] [PubMed] [Google Scholar]
- 34. O'Toole N., Hattori M., Andres C., Iida K., Lurin C., Schmitz-Linneweber C., Sugita M., Small I. (2008) On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25, 1120–1128 [DOI] [PubMed] [Google Scholar]
- 35. Small I. D., Peeters N. (2000) The PPR motif: a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47 [DOI] [PubMed] [Google Scholar]
- 36. Rivals E., Bruyère C., Toffano-Nioche C., Lecharny A. (2006) Formation of the Arabidopsis pentatricopeptide repeat family. Plant Physiol. 141, 825–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmitz-Linneweber C., Small I. (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13, 663–670 [DOI] [PubMed] [Google Scholar]
- 38. Takenaka M., Brennicke A. (2007) RNA editing in plant mitochondria: assays and biochemical approaches. Methods Enzymol. 424, 439–458 [DOI] [PubMed] [Google Scholar]
- 39. Takenaka M., Zehrmann A. (2011) in RNA and DNA Editing (Aphasizhev R., ed) pp. 163–169, Springer, Heidelberg [Google Scholar]
- 40. Takenaka M., Brennicke A. (2009) Multiplex single-base extension typing to identify nuclear genes required for RNA editing in plant organelles. Nucleic Acids Res. 37, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Takenaka M., Brennicke A. (2012) Using multiplex single-base extension typing to screen for mutants defective in RNA editing. Nat. Protoc. 7, 1937–1945 [DOI] [PubMed] [Google Scholar]
- 42. Takenaka M. (2011) in RNA and DNA Editing. (Aphasizhev R., ed) pp. 151–161, Springer, Heidelberg [Google Scholar]
- 43. Clough S. J., Bent A. F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 44. Curtis M. D., Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Knoop V., Rüdinger M. (2010) DYW-type PPR proteins in a heterolobosean protist: plant RNA-editing factors involved in an ancient horizontal gene transfer? FEBS Lett. 584, 4287–4291 [DOI] [PubMed] [Google Scholar]
- 46. Berg M., Rogers R., Muralla R., Meinke D. (2005) Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J. 44, 866–878 [DOI] [PubMed] [Google Scholar]
- 47. Kägi C., Baumann N., Nielsen N., Stierhof Y. D., Gross-Hardt R. (2010) The gametic central cell of Arabidopsis determines the lifespan of adjacent accessory cells. Proc. Natl. Acad. Sci. U.S.A. 107, 22350–22355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmid M., Davison T. S., Henz S. R., Pape U. J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J. U. (2005) A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506 [DOI] [PubMed] [Google Scholar]
- 49. Rüdinger M., Polsakiewicz M., Knoop V. (2008) Organellar RNA editing and plant-specific extensions of pentatricopeptide repeat proteins in jungermanniid but not in marchantiid liverworts. Mol. Biol. Evol. 25, 1405–1414 [DOI] [PubMed] [Google Scholar]
- 50. Rüdinger M., Szövényi P., Rensing S., Knoop V. (2011) Assigning DYW-type PPR proteins to RNA-editing sites in the funariid mosses Physcomitrella patens and Funaria hygrometrica. Plant J. 67, 360–370 [DOI] [PubMed] [Google Scholar]
- 51. Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. (2009) Breaking the code of DNA-binding specificity of TAL-type III effectors. Science 326, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 52. Unseld M., Marienfeld J. R., Brandt P., Brennicke A. (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15, 57–61 [DOI] [PubMed] [Google Scholar]
- 53. Ohtani S., Ichinose M., Tasaki E., Aoki Y., Komura Y., Sugita M. (2010) Targeted gene disruption identifies three PPR-DYW proteins involved in RNA editing for five editing sites of the moss mitochondrial transcripts. Plant Cell Physiol. 51, 1942–1949 [DOI] [PubMed] [Google Scholar]
- 54. Okuda K., Hammani K., Tanz S. K., Peng L., Fukao Y., Myouga F., Motohashi R., Shinozaki K., Small I., Shikanai T. (2010) The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts. Plant J. 61, 339–349 [DOI] [PubMed] [Google Scholar]
- 55. Okuda K., Chateigner-Boutin A. L., Nakamura T., Delannoy E., Sugita M., Myouga F., Motohashi R., Shinozaki K., Small I., Shikanai T. (2009) Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21, 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Okuda K., Shikanai T. (2012) A pentatricopeptide repeat protein acts as a site-specificity factor at multiple RNA-editing sites with unrelated cis-acting elements in plastids. Nucleic Acids Res. 40, 5062–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gobert A., Gutmann B., Taschner A., Gössringer M., Holzmann J., Hartmann R. K., Rossmanith W., Giegé P. (2010) A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 17, 740–744 [DOI] [PubMed] [Google Scholar]
- 58. Khrouchtchova A., Monde R. A., Barkan A. (2012) A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA 18, 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Takenaka M., Zehrmann A., Verbitskiy D., Kugelmann M., Härtel B., Brennicke A. (2012) Multiple organellar RNA-editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5104–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tasaki E., Hattori M., Sugita M. (2010) The moss pentatricopeptide repeat protein with a DYW domain is responsible for RNA editing of mitochondrial ccmFc transcript. Plant J. 62, 560–570 [DOI] [PubMed] [Google Scholar]
- 61. Grewe F., Herres S., Viehöver P., Polsakiewicz M., Weisshaar B., Knoop V. (2011) A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 39, 2890–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]