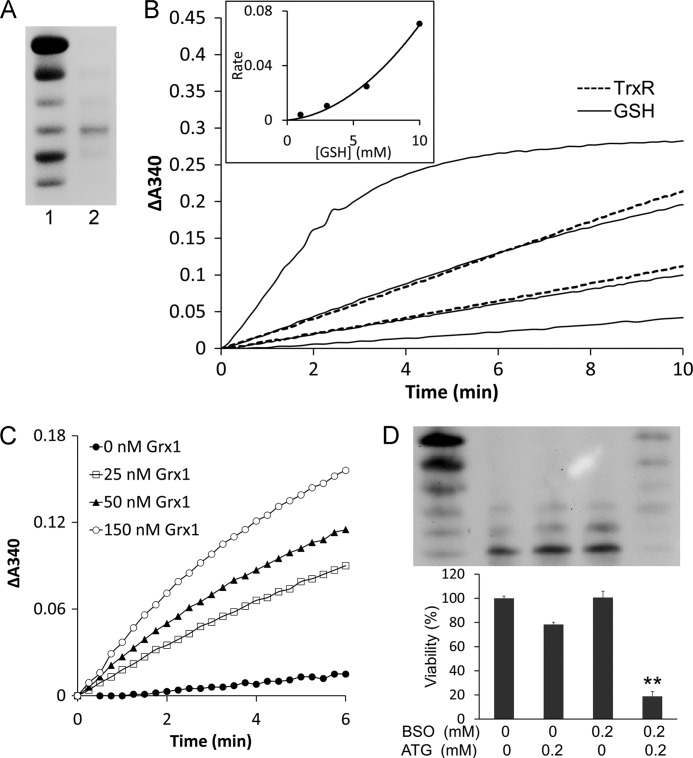
FIGURE 7.
The glutathione system was a backup of TrxR1for reduction of Trx1. A, Trx1-S2 was prepared as described under “Experimental Procedures” and confirmed by redox urea-PAGE. Lane 1, mobility standard. Lane 2, Trx1-S2. B, 60 nm glutathione reductase, 0.2 mm NADPH, and 50 μm Trx1-S2 were added to cuvettes for the GSH reduction assay in the presence of 1, 3, 6, and 10 mm GSH (four solid lines from bottom to top). 0.2 mm NADPH and 50 μm Trx1-S2 were added to cuvettes for the TrxR1 reduction assay in the presence of 5 and 10 nm TrxR1 (two dotted lines from bottom to top). The absorbance at 340 nm was followed against an identical blank without Trx1-S2. A second-order polynomial regression line was fit in the inset. C, 60 nm glutathione reductase, 0.2 mm NADPH, 1 mm GSH, indicated amounts of Grx1 and 50 μm Trx1-S2 was added to cuvettes. The absorbance at 340 nm was followed against an identical blank without Trx1-S2. D, HeLa cells were treated with the indicated concentrations of ATG and/or BSO. After 48-h treatment, the redox state of Trx1 was detected using a redox Western blot analysis. The cell viability was measured with an MTT assay. Error bars show mean ± S.D. n = 4. **, p < 0.01, Student's t test, treated cells versus control.