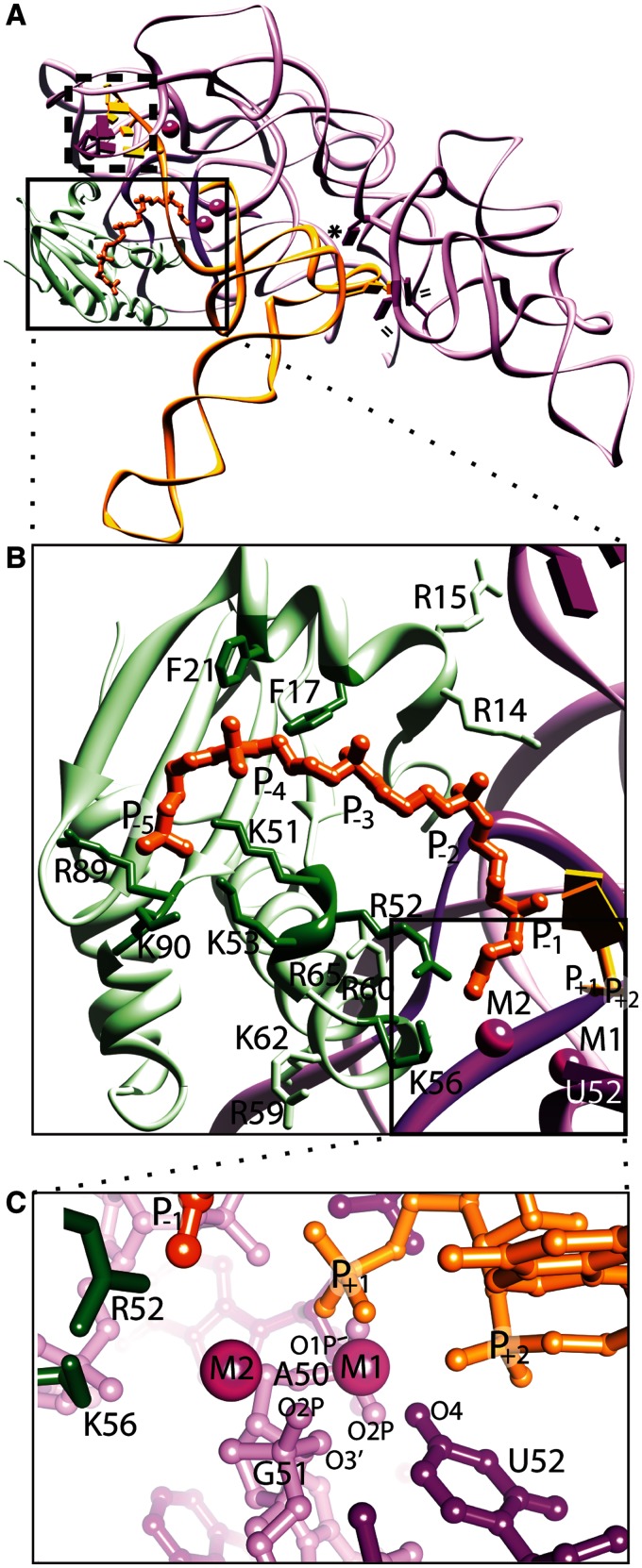
Figure 1.
Structure of bacterial RNase P holoenzyme in complex with tRNA and a short oligonucleotide leader (rcsb: 3Q1R). The T. maritima holoenzyme consists of a large P RNA (purple), a small protein (light green), and critical metal ions (magenta spheres), and makes several contacts with the tRNA (orange) and a short leader oligonucleotide (dark orange). (A) Ribbon representation emphasizing the tertiary P RNA-tRNA elements crucial for recognition, where double lines (=) represent hydrophobic base stacking, an asterisk (*) represents an A-minor interaction, and a dashed box represents intermolecular base pairs. In the structure of the complex, the protein sits atop and stabilizes conserved, non-helical regions of the P RNA (dark purple). The boxed region highlights the area where the P protein, P RNA and tRNA converge, including the active site. (B) Expanded view of the boxed region in (A), showing key protein residues that contact the P RNA (light green, including R14, R15, R59, R60, K62, R65), or residues that potentially contact the pre-tRNA leader region (dark green, including F17, F21, K51, R52, K53, K56, R89, K90). The phosphates of the pre-tRNA leader (labelled P−1 – P−5, dark orange) were observed in a structure of a holoenzyme-tRNA complex with a short leader soaked into the crystal, and the active site region (boxed) was inferred from the location of the +1 phosphate of the tRNA and the presence of two catalytically important metal ions (M1 and M2). (C) An expanded view of the active site of the bacterial RNase P enzyme. Specific P RNA oxygen atoms, including: G51 (O2P and O3′), A50 (O1P and O2P) and the O4 atom of a universal uridine nucleobase (U52), are likely to make contacts with the metal ions. Protein residues K56 and R52 are located ∼4.5 Å from the nearest metal ion (M2) and ∼7–8 Å from the phosphorus atom of the mature tRNA (+1 nucleotide). All structural figures were prepared using Chimera (61).