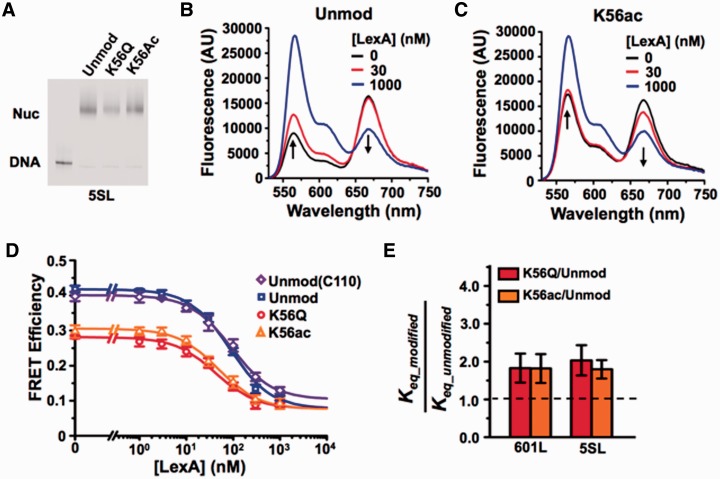
Figure 2.
DNA sequence does not influence H3(K56ac) enhancement of TF binding. (A) Cy3 fluorescence image of native PAGE analysis of purified FRET-labeled nucleosomes containing the 5SL NPS and unmodified H3 (lane 2), H3(K56Q) (lane 3) or H3(K56ac) (lane 4). (B and C) Fluoresence emission spectra of 5SL FRET-labeled nucleosomes containing unmodified H3 or H3(K56ac), respectively, excited at 510 nm (donor excitation) in the presence of 0.5× TE, 1 mM NaCl and LexA at 0 nM (black), 30 nM (red) or 1000 nM (blue), (D) Steady-state FRET efficiency, as determined by the (ratio)A method (36), versus LexA concentration for nucleosomes containing unmodified H3 (blue), H3(C110) (purple), H3(K56Q) (red), and H3(K56a) (orange) at 1 mM NaCl. Plots are the average of three LexA titrations and the error bars were determined from the SD of the three measurements. The data were fit to a non-cooperative binding curve, which determines S0.5-nuc, the LexA concentration at which 50% of the nucleosomes are bound by LexA. (E) Relative change in Keq for nucleosomes with the 601L (14) or 5SL NPS containing H3(K56Q) or H3(K56ac) versus unmodified H3 at 1 mM NaCl. This is inversely related to the relative change in S0.5.