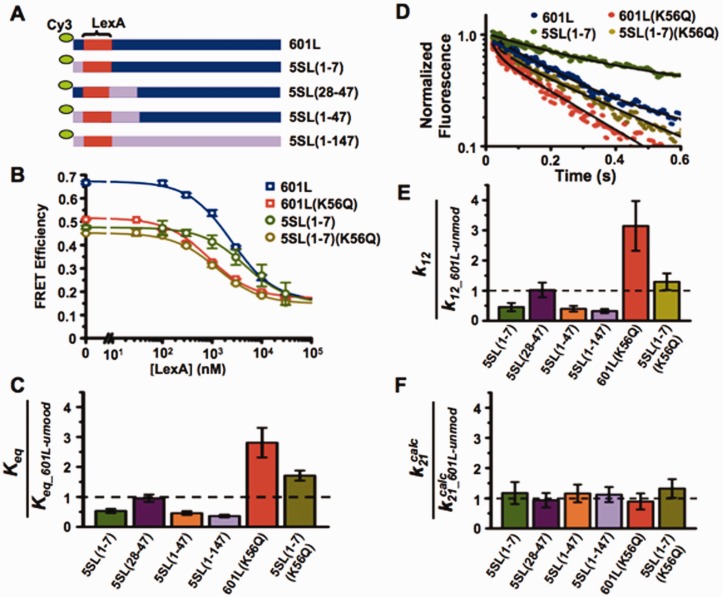
Figure 3.
DNA sequence between the nucleosome entry–exit and the TF-binding site impacts nucleosome unwrapping. (A) DNA constructs with the 601 sequence (blue) iteratively replaced by the 5S sequence (lavender), where the parenthetical numbers represents the bases in 601 replaced by 5S; lexA site in red. (B) Steady-state FRET efficiency, as determined by the (ratio)A method, versus lexA concentration for nucleosomes containing 601L and unmodified H3 (blue), 601L and H3(K56Q) (red), 5SL(1–7) and unmodified H3 (green), and 5SL(1–7) and H3(K56Q) (olive) at 75 mM NaCl. Plots are the average of three LexA titrations and the error bars were determined from the SD of the three measurements. The data were fit to a non-cooperative binding curve. (C) Relative Keq for the indicated nucleosome versus nucleosomes containing 601L and unmodified H3 at 75 mM NaCl. (D) Normalized stopped flow Cy5 emission versus time at 15 μM LexA for nucleosomes containing 601L and unmodified H3 (blue), 601L and H3(K56Q) (red), 5SL(1–7) and unmodified H3 (green), and 5SL(1–7) and H3(K56Q) (olive) at 75 mM NaCl. (E and F) Relative unwrapping and calculated rewrapping rates, respectively, for the indicated nucleosomes versus nucleosome containing 601L and unmodified H3 at 75 mM NaCl (see also Supplementary Figure S4).