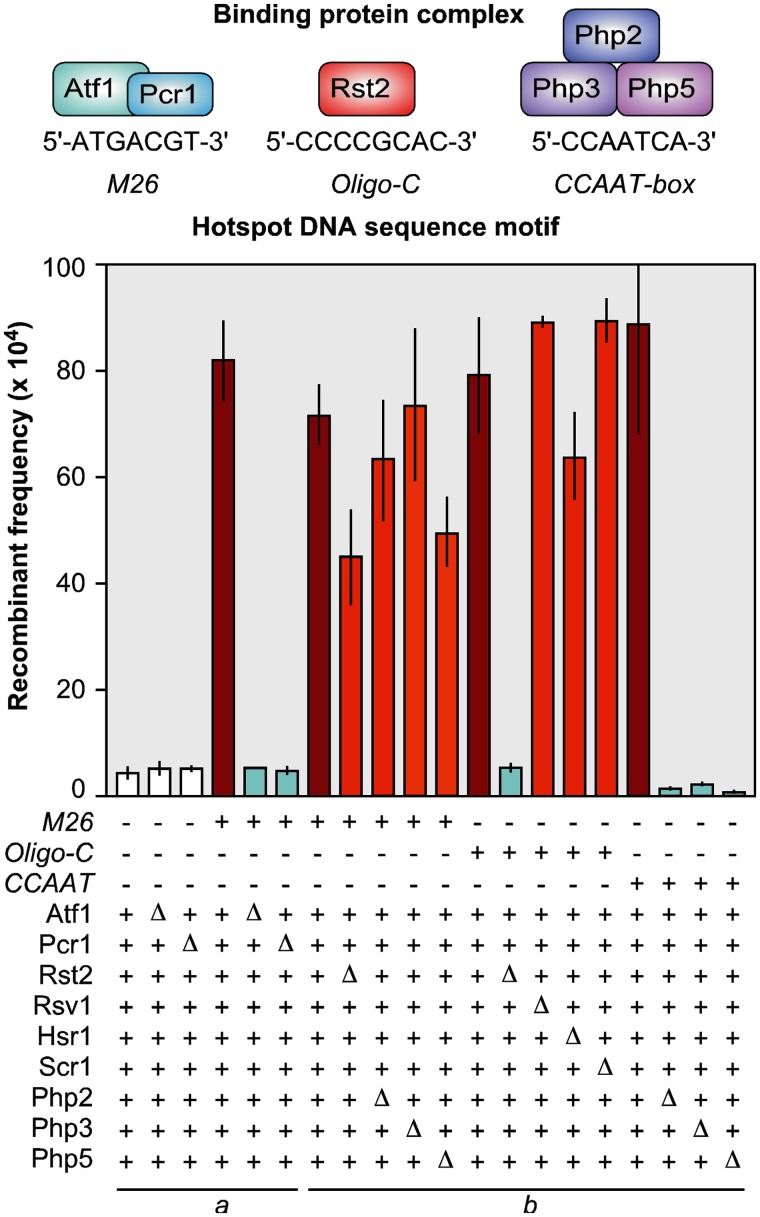
Figure 2.
Discrete protein–DNA complexes are essential for hotspot activity, act in cis with high specificity, and can function redundantly. Three hotspot motifs have been defined experimentally at high resolution (e.g. Figure 1) and have binding-protein complexes whose hotspot-regulating functions have been characterized by mutation (5–7,12,32). Top panel depicts protein complexes of fission yeast (orthologs are present in other taxa) and DNA sites to which they bind. Plotted are the effects of each DNA site and binding protein upon recombinant frequencies at the same test locus. Values for M26 are presented twice to match data set comparisons in the original articles (underscored groups). Note three things. First, each DNA site generates a hotspot (red) relative to negative controls (white). Second, for each DNA site-dependent hotspot, the corresponding binding proteins are essential for hotspot activity (light blue). Third, the proteins that bind to and promote recombination at their cognate DNA site have no significant or only modest effects upon hotspot activity of other DNA sites (orange). Similarly, zinc-finger proteins Rsv1, Hsr1 and Scr1 have no significant effect upon Oligo-C hotspot activity, which is conferred by binding of zinc-finger protein Rst2. Plotted from data in a(32) © 1997 and b(7) © 2011 with permission from the National Academy of Sciences and the Genetics Society of America.