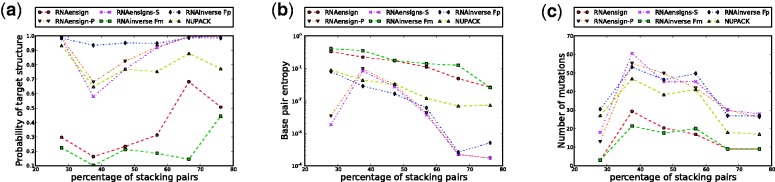
Figure 4.
Comparison of the probability optimized RNA-ensign (blue line) with NUPACK (green line) and the original version of RNA-ensign (red line). This benchmark has been realized on 100 secondary structure targets of length 60 with 10 random seeds for each target. The x-axis represents the number of stacks in the target structure. In the leftmost figure (a), the y-axis represents the probability go the target structure on the sequence. In the middle figure (b), the y-axis indicates the entropy of the solutions using a log-scale. In the rightmost figure (c), the y-axis reports the number of mutations between the seed and the solution.