Table 1.
Re-engineering of riboswitches
| Target | RNAinverse |
RNAensign |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 100K |
310K |
1000K |
|||||||||
| length | 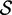 |
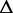 G G |
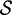 |
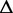 G G |
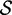 |
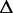 G G |
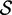 |
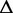 G G |
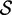 |
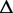 G G |
|
| Ade ydhL gene OFF | 110 | 0.238 | −17.2 | 0.118 | −25.0 | 0.169 | −25.5 | 0.100 | −25.7 | 0.116 | −24.7 |
| Ade ydhL gene ON | 110 | 0.238 | 17.2 | 0.237 | −6.1 | 0.343 | −9.2 | 0.352 | −3.7 | ||
| Ade add gene OFF | 113 | 0.446 | −1.8 | 0.253 | −5.8 | 0.075 | −9.2 | 0.073 | −15.2 | 0.088 | −8.8 |
| Ade add gene ON | 113 | 0.446 | 1.8 | 0.307 | −9.8 | 0.159 | −9.8 | 0.189 | −10.8 | ||
| c-di-GMP OFF | 124 | 0.381 | −8.8 | 0.237 | −25.3 | 0.233 | −27.7 | 0.225 | −38.0 | 0.304 | −27.3 |
| c-di-GMP ON | 124 | 0.381 | 8.8 | 0.292 | −15.1 | ||||||
| SAM OFF | 134 | 0.302 | −15.3 | 0.123 | −15.2 | 0.213 | −29.0 | 0.183 | −22.7 | 0.147 | −22.5 |
| SAM ON | 134 | 0.302 | 15.3 | ||||||||
| xpt-pubX OFF | 148 | 0.073 | −18.3 | 0.101 | −22.7 | 0.116 | −31.6 | 0.117 | −18.9 | ||
| xpt-pubX ON | 148 | 0.073 | 18.3 | 0.435 | −5.8 | ||||||
Re-engineering of riboswitches using RNAinverse (we report the best results among 1000 runs) and RNA-ensign. Only solutions with less than 10% of mutations have been considered. We selected the solution with the lowest entropy. For each method, we report the entropy 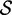 and the difference of energies
and the difference of energies 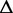 G between the two conformation ‘ON’ and ‘OFF’ of the riboswitches. To illustrate the versatility of our approach, we ran RNA-ensign with three different formal temperatures: T = 100 (increased thermodynamic pressure), T = 310 (default) and T = 1000 (reduced thermodynamic pressure).
G between the two conformation ‘ON’ and ‘OFF’ of the riboswitches. To illustrate the versatility of our approach, we ran RNA-ensign with three different formal temperatures: T = 100 (increased thermodynamic pressure), T = 310 (default) and T = 1000 (reduced thermodynamic pressure).
